STR molecule marker based on sika deer whole genome developing and application of STR molecule marker
A molecular marker, whole-genome technology, applied in recombinant DNA technology, microbial determination/inspection, DNA/RNA fragments, etc., to reduce costs, improve accuracy, and facilitate genotyping
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
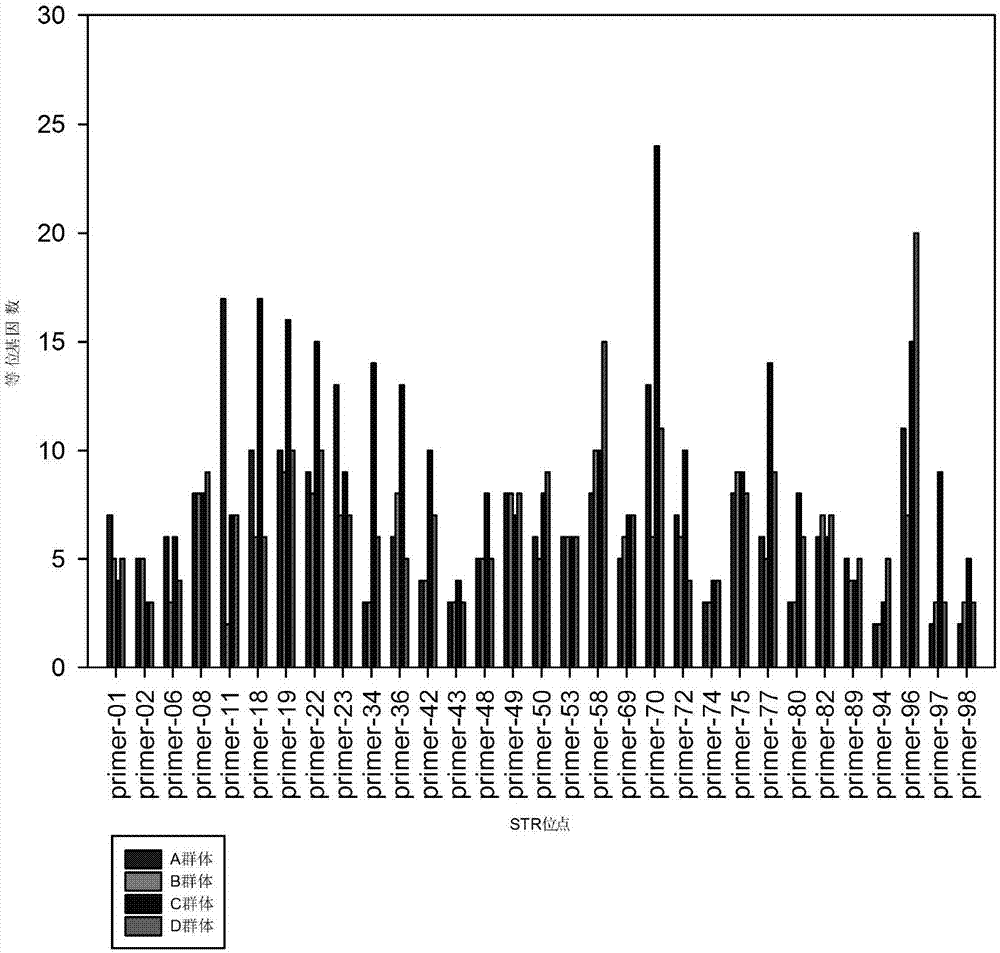
[0072] Example 1 STR site screening and polymorphism analysis of 4-base repeat unit in sika deer
[0073] According to the results of sika deer genome sequencing and sika deer population resequencing, MISA analysis was first used to obtain 940,413 STR sites in the reference genome, and 14,863 STR sites containing 4-base repeating units were extracted; then searched the database for STR site polymorphisms There are 386 STR intervals containing InDels with a screening threshold greater than 7000.
[0074] A two-step method can be used to quickly screen the STR sites of highly polymorphic 4-base repeat units in the sika deer genome.
Embodiment 2
[0075] Example 2 Batch design of STR primers
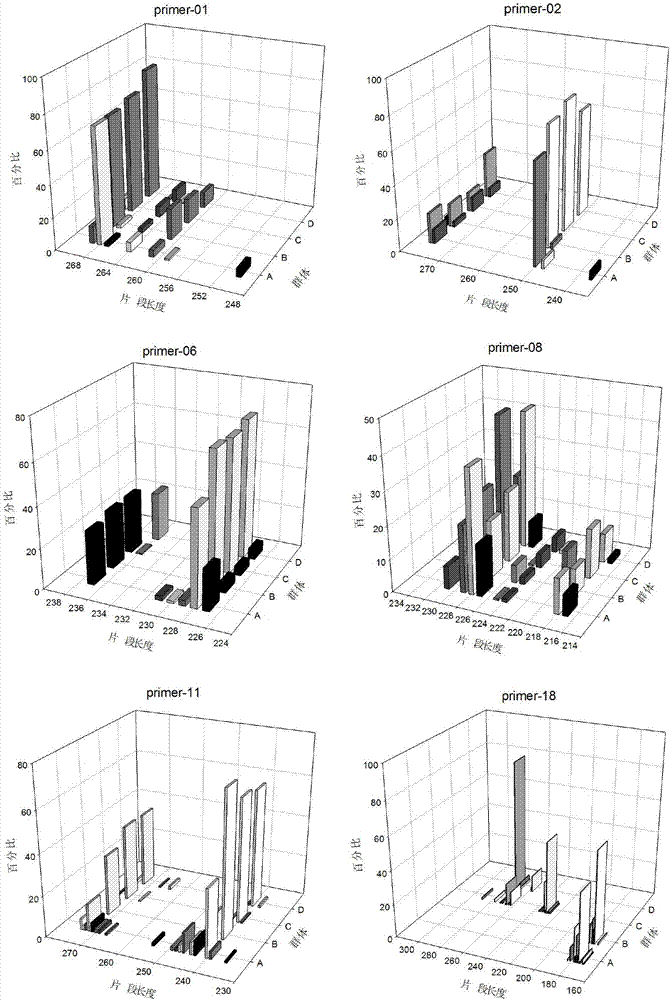
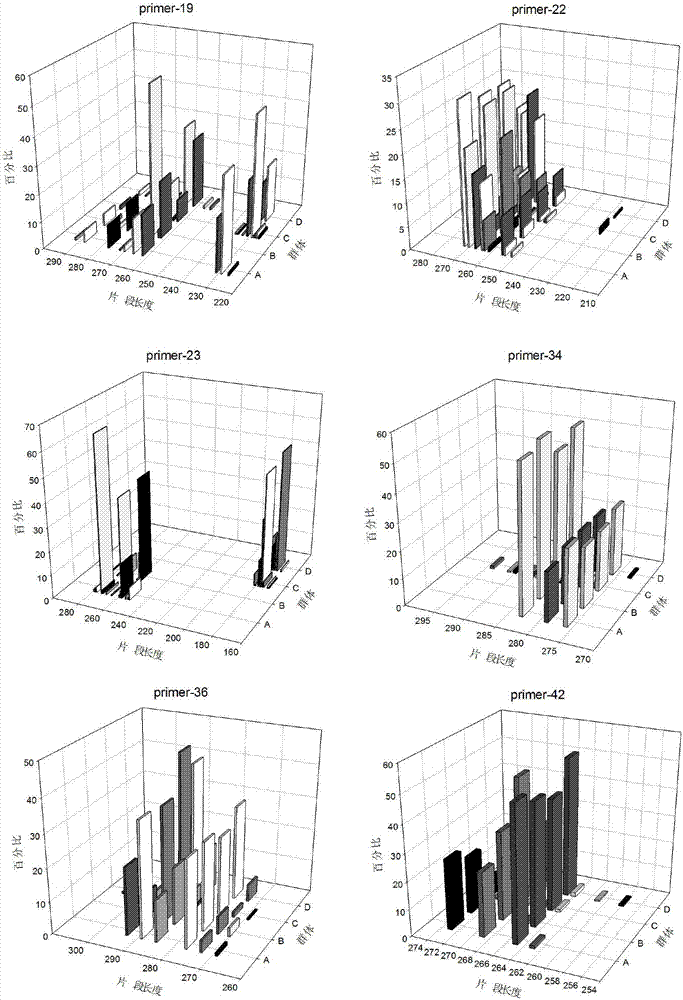
[0076] Use Primer3 to design primers in batches for the screened STR sites. The primer length is limited to 20bp±2bp, the annealing temperature is controlled between 55°C-60°C, and the CG% is controlled between 30%-80%. Elimination is prone to mismatches And primers with hairpin structure, the product fragment size is controlled between 80-300bp. 284 pairs of STR primers were designed for 386 high-length polymorphic sites, and e-PCR was performed with the designed primers to remove non-specific amplification. Through comparative analysis in NCBI, try to make the screened STR markers cover all chromosomes , to avoid being located at the end of the chromosome, select 100 pairs of primers for subsequent polymorphism verification.
[0077] The Primer3 was used to design primers in batches, and the designed primers were screened and verified by e-PCR, and further screened by sequence comparison in the NCBI database. The developm...
Embodiment 3
[0078] Example 3 Polymorphism and authenticity verification of STR loci
[0079] The DNA of 8 unrelated sika deer individuals was extracted, and the extracted sika deer genomic DNA was used as a template for PCR amplification, and the primers were synthesized by Shanghai Sangong Biotechnology Company. A polyacrylamide gel was prepared, the amplified products were detected by electrophoresis and silver staining, and 31 pairs of primers with high polymorphism were screened according to the electrophoresis bands for subsequent sequencing verification. The PCR products amplified by 31 pairs of primers were sequenced, compared with the e-PCR amplification sequence using the reference genome as a template, and through consistency analysis, it was determined that all 31 STR sites were real polymorphic sites .
[0080] Through polyacrylamide gel detection and sequencing analysis, all 31 STR sites were real polymorphic sites.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com