Paphiopedilum Henryanum EST-SSR (expressed sequence tag-simple sequence repeat) marker primers and their development method and application
A Paphiopedilum and labeling technology, which is applied in biochemical equipment and methods, microbiological determination/inspection, DNA/RNA fragments, etc., can solve problems that have not been reported, and achieve good repeatability, low production cost, and polymorphism. sexual enrichment effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1. Development of P. henrypipes EST-SSR primers
[0051] 1. EST-SSR primer design
[0052] Experimental materials: P. henrylis organs were obtained from the greenhouse of the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences; various reagents or medium components used in all the examples of the present invention were obtained commercially.
[0053] 1) Construction of transcriptome library: Extract total RNA from Papudendron henrylium organs, isolate mRNA, synthesize and purify cDNA by reverse transcription, end repair, add adenine nucleosides to connect sequencing adapters, and recover the size of 200-700bp by agarose gel electrophoresis The recovered fragments were amplified by PCR to construct a transcriptome library.
[0054] 2) Acquisition of transcriptome data: Sequencing the transcriptome library obtained in step 1) to obtain transcriptome sequencing data, and splicing the sequencing data to obtain sequence data of unigenes. ...
Embodiment 2
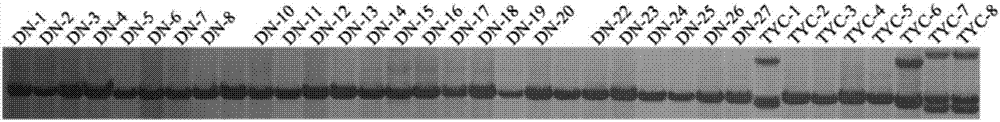
[0075] Example 2. Analysis of Genetic Diversity Using P. henrypipes EST-SSR Primers
[0076] Experimental materials: The experimental samples of P. henryi were obtained from 8 plants of Taiyang Chong population (TYC) and 25 plants of Dalong population (DN) in Malipo County, Wenshan Prefecture, Yunnan Province.
[0077] Analysis of the amplified polymorphism information of these 34 pairs of EST-SSR primers (see Table 3 below) shows that the observed and expected heterozygosity ranges are 0.0303-1.000 and 0.0597-0.6793, respectively, and the distribution range of the Shannon information index is 0.14- 1.25, with an average value of 0.61, indicating that the genetic differentiation of these 33 P. henryii materials is relatively rich.
[0078] Table 3 Analysis of polymorphism information amplified by 34 pairs of EST-SSR primers
[0079]
[0080]
[0081] In addition, these 34 pairs of primers were also used to perform cluster analysis on 33 different P. henley species from ...
Embodiment 3
[0082] Example 3. Generic analysis in Paphiopedilum using EST-SSR primers
[0083] The generality analysis of other six species of P. genus from the genus Paphiopedilum was carried out using microsatellites of P. henley. The results (see Table 4 below) showed that 34 pairs of EST-SSR primers were used in P. villosum, The successful expansion rate of P. venustum, P. wardii, P. dianthum, P. concolor and P. micranthum are 0.85, 0.82, 0.74, 0.68, 0.56, and 0.56, respectively, indicating that these 34 pairs of EST-SSR primers have high versatility in the genus Paphiopedilum, especially in the subgenus Paphiopedilum, which shows that The closer the genetic distance to P. henlensis, the higher the amplification efficiency of SSR primers. It was proved that 34 pairs of EST-SSR primers developed by using the transcriptome sequencing of P. henrifolia can be used for the study of genetic diversity and genetic structure of closely related species.
[0084] Table 4 Amplification of 34 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com