Fluorescence chemical sensor for detecting DNA adenine transmethylase, and detection method thereof
A fluorescent chemical sensing and methyltransferase technology, applied in the field of biological analysis, can solve problems such as the complexity of experimental operations that increase the difficulty of program design, the limited application of electrode surface modification, and the fast response speed of electrochemical methods, to prevent The effect of non-specific reaction, reduced experimental cost, simple preparation and detection method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
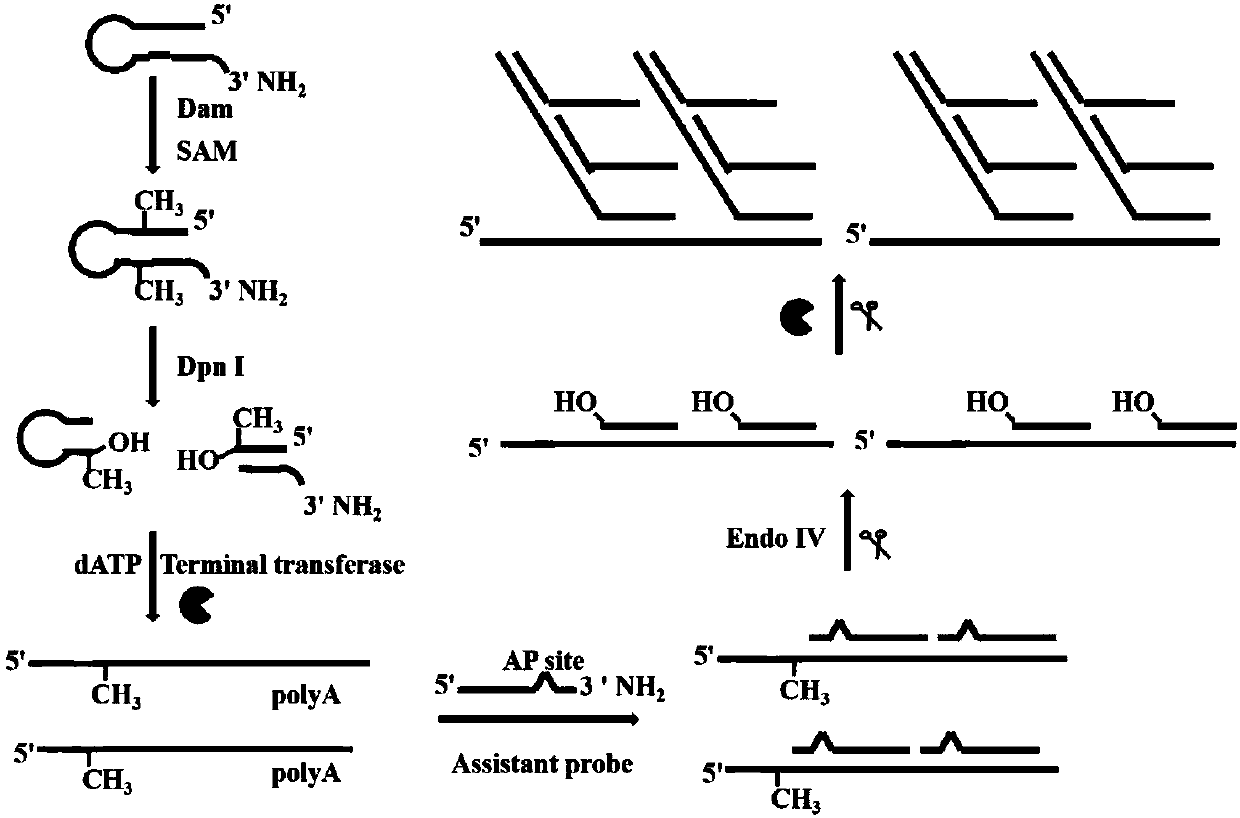
[0069] Experimental method steps
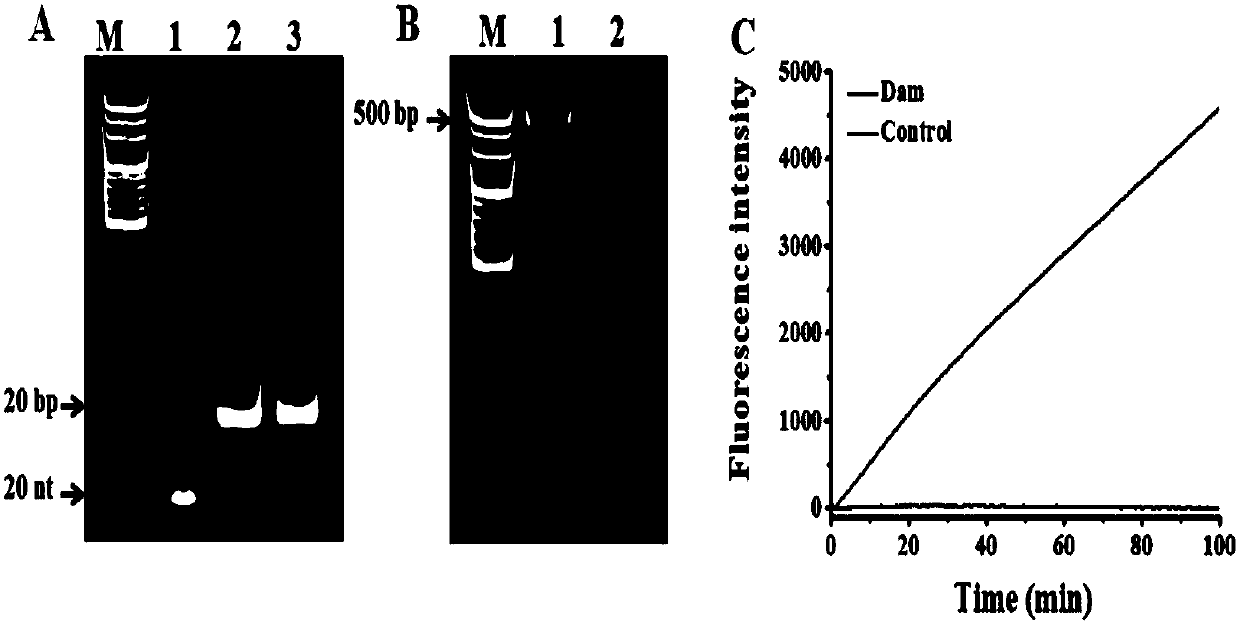
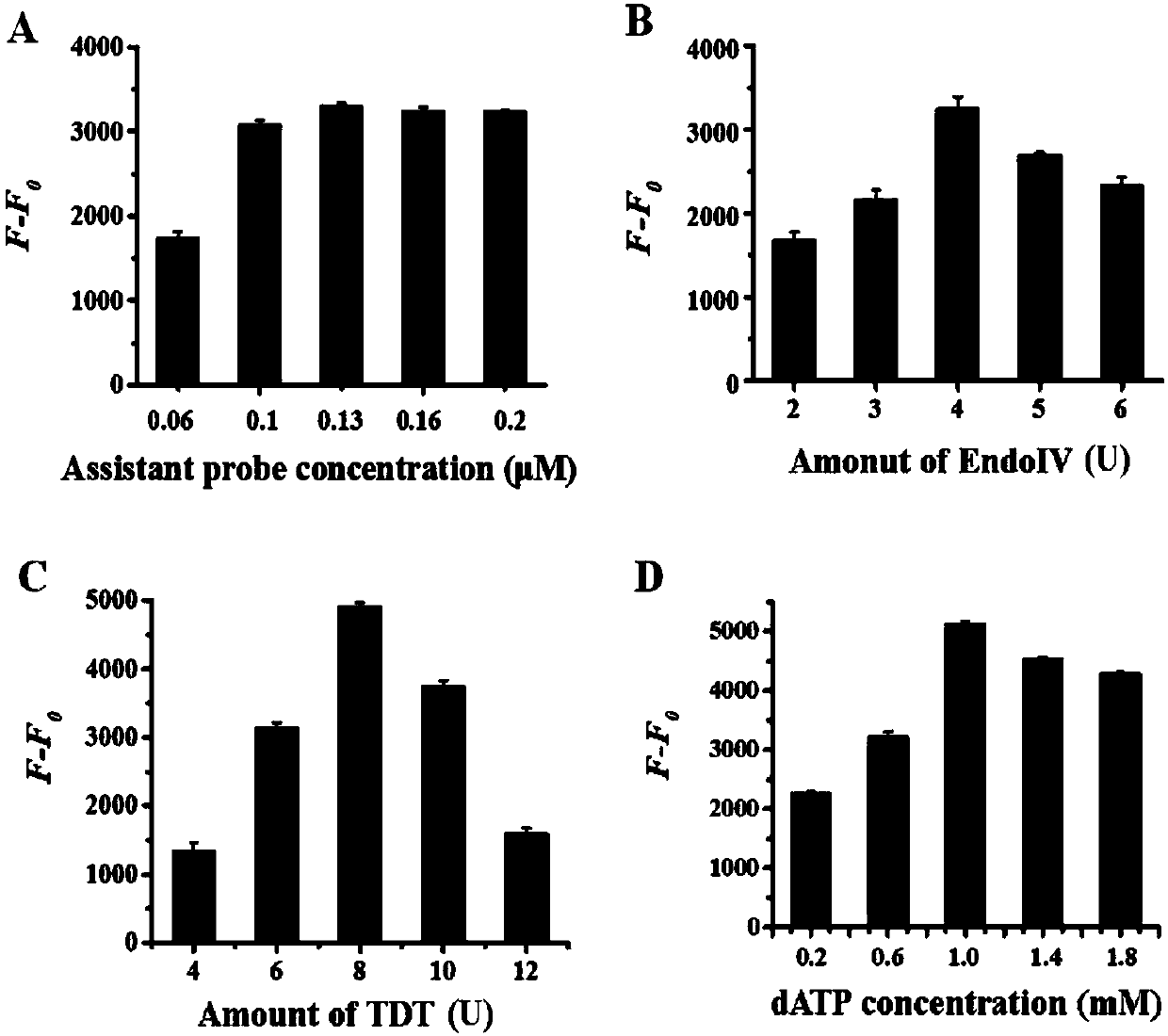
[0070] 1. Detection of Dam methyltransferase: in the hairpin probe containing 0.5 micromole per liter, 160 micromole per liter SAM (S-adenosylmethionine), 10 units of restriction endonuclease DpnI, 1 × 200 μl reactions of DNADam buffer (50 mmol / L Tris-HCl buffer, 10 mmol / L EDTA, 5 mmol / L 2-mercaptoethanol, pH 7.5) and different amounts of Dam methyltransferase Methylation and cleavage of the hairpin probes are performed in the mixture. The mixture was incubated at 37°C for 2 hours and then inactivated at 80°C for 20 minutes. In 30 μl of 1× terminal transferase buffer (50 mmol / L potassium acetate, 20 mmol / L Tris-Ac, 10 mmol / L magnesium acetate, pH 7.9), 0.25 mmol / L dichloride Cobalt, 1×SYBR Gold, 0.13 micromole per liter of auxiliary probe reaction solution for hyperbranched amplification, 1 millimole per liter of dATPs (deoxyadenosine triphosphate), 4 units of EndoⅣ (endonuclease IV), 8 Unit TDT (terminal transferase) and 4 microliters of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com