Single-nucleotide polymorphism marker locus, primer pair and kit for identifying single-petal/double-petal traits of peach blossoms and application
A technology of single nucleotide polymorphism and marker loci, applied in the biological field, can solve the problems of long distance, low accuracy, and small number of genomes of target trait genes, and achieve good social and economic benefits and low cost effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] The acquisition of embodiment 1 SNPs marker locus
[0027] In the present invention, 129 peach germplasms randomly obtained from the peach germplasm resource garden of Zhengzhou Institute of Pomology, Chinese Academy of Agricultural Sciences were used as samples, and the sample DNA was extracted by conventional CTAB method, and the 129 peach germplasms were reconstructed by Illumina HiSeq 2000 sequencer. Sequencing obtained 121Gb data, covering an average of 89.28% of the peach genome, and the average sequencing depth was about 4.21×. According to the 50-150bp reads obtained by sequencing, compared with the peach reference genome version 2 (http: / / www.rosaceae.org / node / 355), 4,063,377 SNPs were identified, and these SNPs were used to analyze 129 germplasm Genome-wide association analysis was performed on the flower type (single / double) traits, and the SNP that was significantly associated with peach flower type (single / double) was identified at the 25,058,942 bp of chro...
Embodiment 2
[0028] Embodiment 2 Utilizes the method for identifying peach flower type (single petal / double petal) character with SNP marker
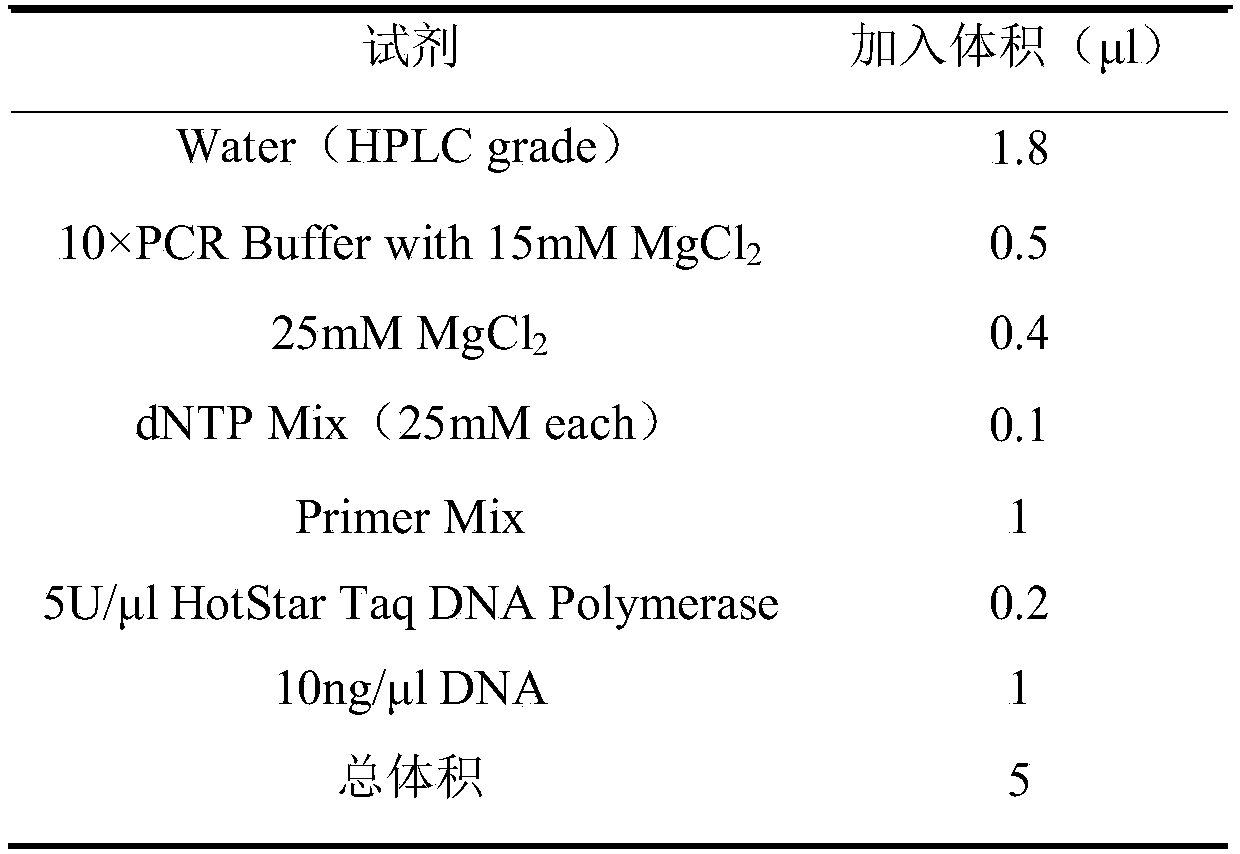
[0029] 1. DNA extraction
[0030] The DNA of the peach sample tissue to be tested was extracted by the conventional CTAB method, and the RNA was removed. The total volume of the DNA sample was not less than 15 μl. Measure the OD value of the DNA sample at 260nm and 280nm with a UV photometer, and calculate the DNA content and OD 260 / 280 ratio. DNA sample purity OD 260 / 280 The value should be between 1.8-2.0 and the concentration should be diluted to 10ng / μl.
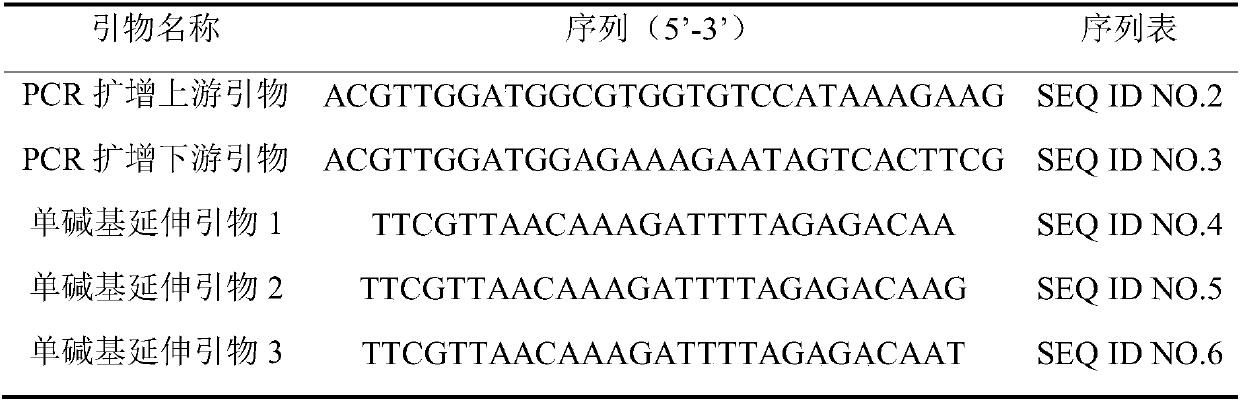
[0031] 2. Design primers
[0032] Primers were designed according to the 150 bp sequences at positions 25, 058, and 942 of chromosome 2 of the peach genome version 2 (see Table 1 for specific nucleotide sequences).
[0033] Table 1 SNP flanking sequence information
[0034]
[0035] Among them, K represents G or T.
[0036] After the primers were synthesized by the biotechnology comp...
Embodiment 3
[0063] Example 3 337 peach natural populations use flower type (single petal / double petal) associated SNP markers to carry out blind test verification of phenotypic traits 1. Selection of experimental materials
[0064] 337 natural populations of common peach germplasm preserved in the resource garden of Zhengzhou Pomology Research Institute were used as experimental materials, including 16 double petals and 321 single petals.
[0065] 2. Identification method using peach-blossom-associated SNP markers
[0066] Using positions 25,058,942 of the second chromosome of the peach genome of the present invention as the nucleotide polymorphism marker site, 337 germplasms were blindly tested and identified. For the specific identification method, refer to the method in Example 2.
[0067] 3. The predictive ability of typing results to phenotype in natural populations (Table 6)
[0068] From the identification results, it can be seen that among the 321 single lobes, only 1 was typed a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com