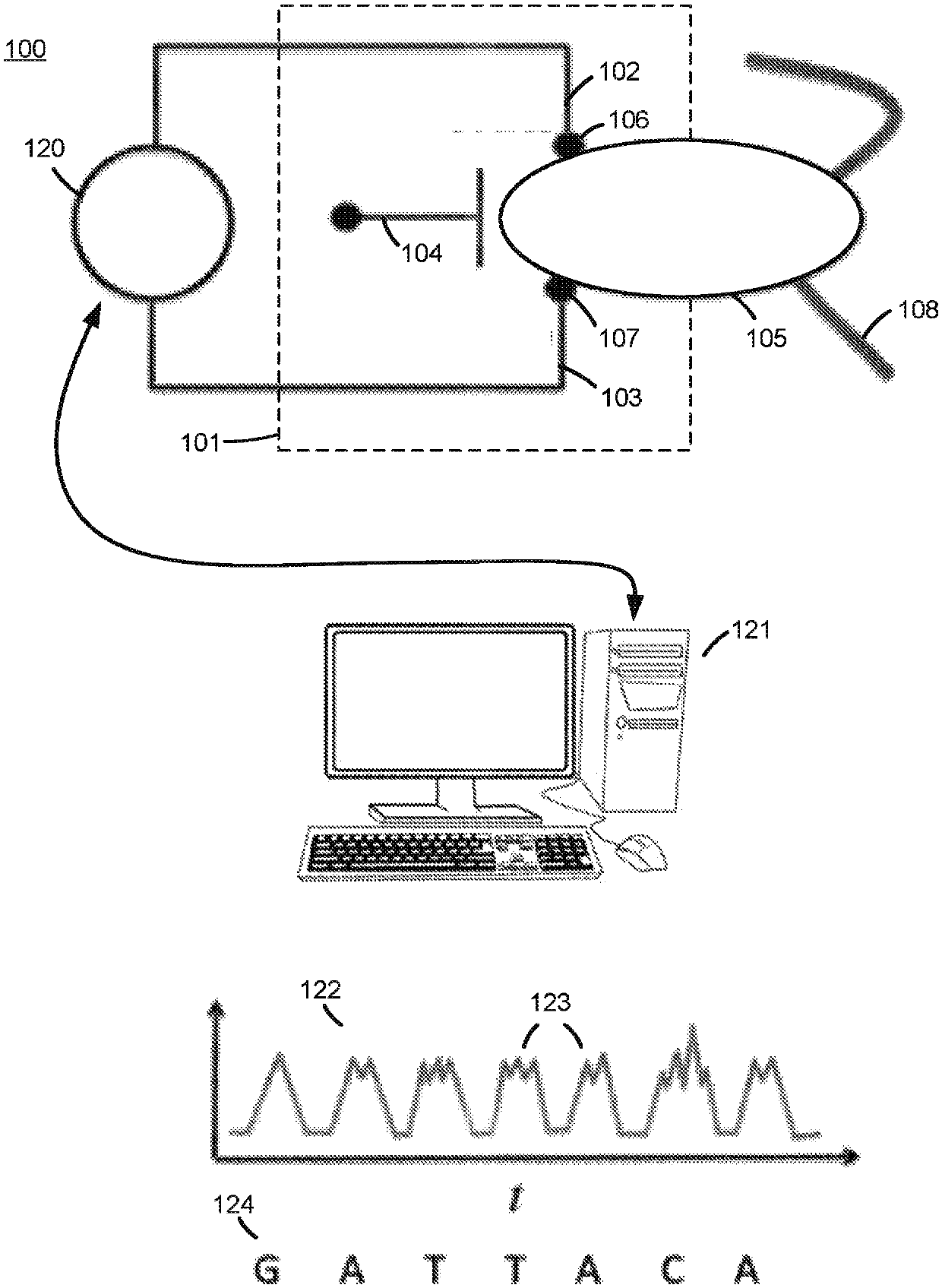
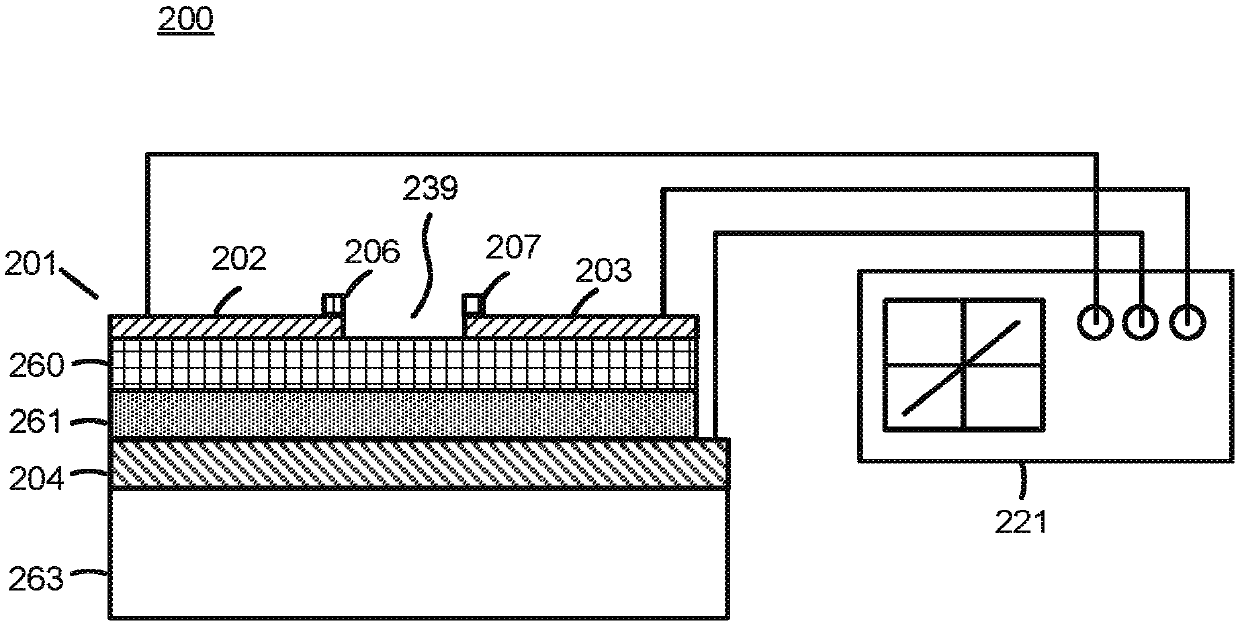
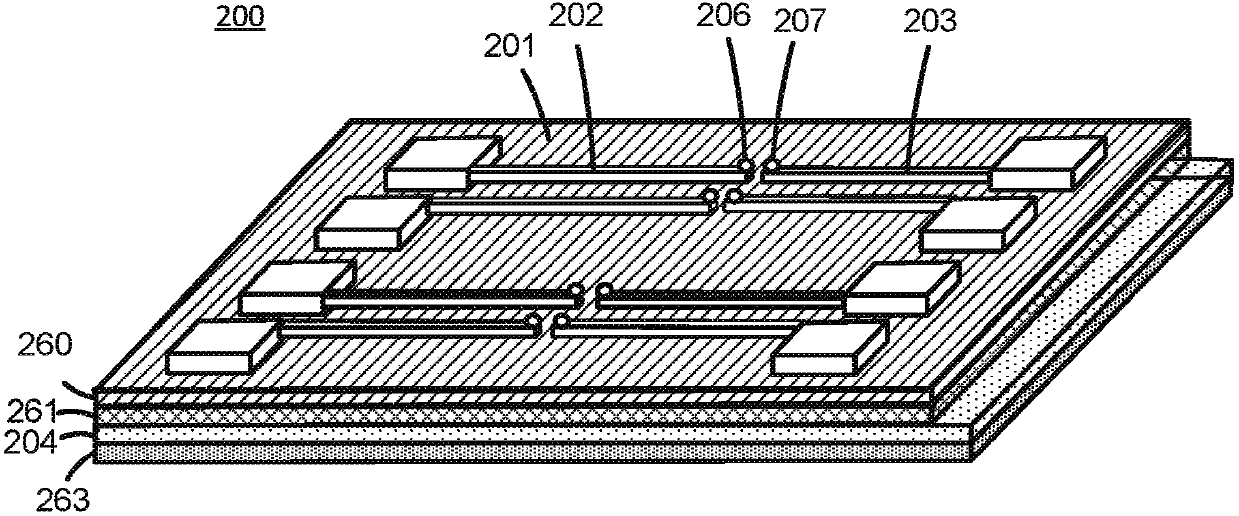
Biomolecular sensors and methods
A sensor and molecular technology, applied in biochemical equipment and methods, nanotechnology for sensing, and microbial assay/inspection, etc., which can solve the functional barriers, inability to engineer design, sensitivity and Issues such as sensor density limitations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0103] Self-assembly of biopolymer bridges
[0104] The end-to-end length was constructed using an oligonucleotide of SEQ ID NO: 1 containing a 5'-thiol modification and an oligonucleotide of SEQ ID NO: 2 containing a 5'-thiol modification and an internal biotin modification A double-stranded DNA biopolymer bridge molecule of approximately 20 nm. Bridge molecules were labeled for visualization purposes using a streptavidin-gold tag. A test array 1200 of gold nanoparticle contacts was fabricated using electron beam lithography ( Figure 12 ) to deposit pairs of gold contacts, each pair of contacts defining a center-to-center contact gap of about 20 nm. A buffer solution containing gold-labeled bridging molecules was contacted with the test array of gold nanoparticle contacts. After a brief incubation period, excess solution was removed, the arrays were washed and imaged by scanning electron microscopy (SEM). Figure 12 A SEM image showing the arrangement of gold contacts 12...
Embodiment 2
[0106] Detection of self-assembly steps
[0107] A sensor device with a single sensor comprising gold contacts attached to electrodes with a center-to-center contact gap of about 20 nm was fabricated using electron beam lithography. The sensor is packaged with a PDMS flow cell that includes a 1 mm wide by 0.4 mm high channel that is open at either end to allow the introduction of liquid into the first end inside the flow cell and the discharge of liquid from the second end of the flow cell. , and the pool contacts the sensor solution. The direction of the flow cell channel is normal to the direction of the electrodes containing the sensor, which is located approximately halfway along the length of the flow cell channel. A low ionic strength buffer solution was introduced into the flow cell and a 0.5 V potential was applied to the sensor throughout the following successive steps: Introduction and self-assembly of double-stranded DNA bridge molecules (as described above in Exam...
Embodiment 3
[0114] Detection of nucleotide base incorporation
[0115] Sensor devices comprising biopolymer bridge molecules and Klenow fragment probes were fabricated and assembled as described in Example 2 above. A sensor device is used to generate signal traces in response to DNA synthesis reactions performed using single-stranded DNA templates of varying length and sequence composition. Figure 17 Signal traces for providing template sequences incorporating single bases are shown. The signal feature at 0.5 s was interpreted as corresponding to template-dependent activity and base incorporation of the Klenow fragment, and the much weaker signal feature after 0.6 s was interpreted as corresponding to some form of noise or spuriousness in the system. Signal. Figure 18 Signal traces for various template regions are shown. The template and primers described in Example 2 above were used in the reactions shown.
[0116] The top and bottom signal traces are control experiments where dNTP...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com