A set of maize chloroplast InDel markers applicable to capillary electrophoresis detection platform
A chloroplast and corn technology, applied in the field of crop molecular biology, can solve problems such as unrecorded chloroplast genome polymorphic sites
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
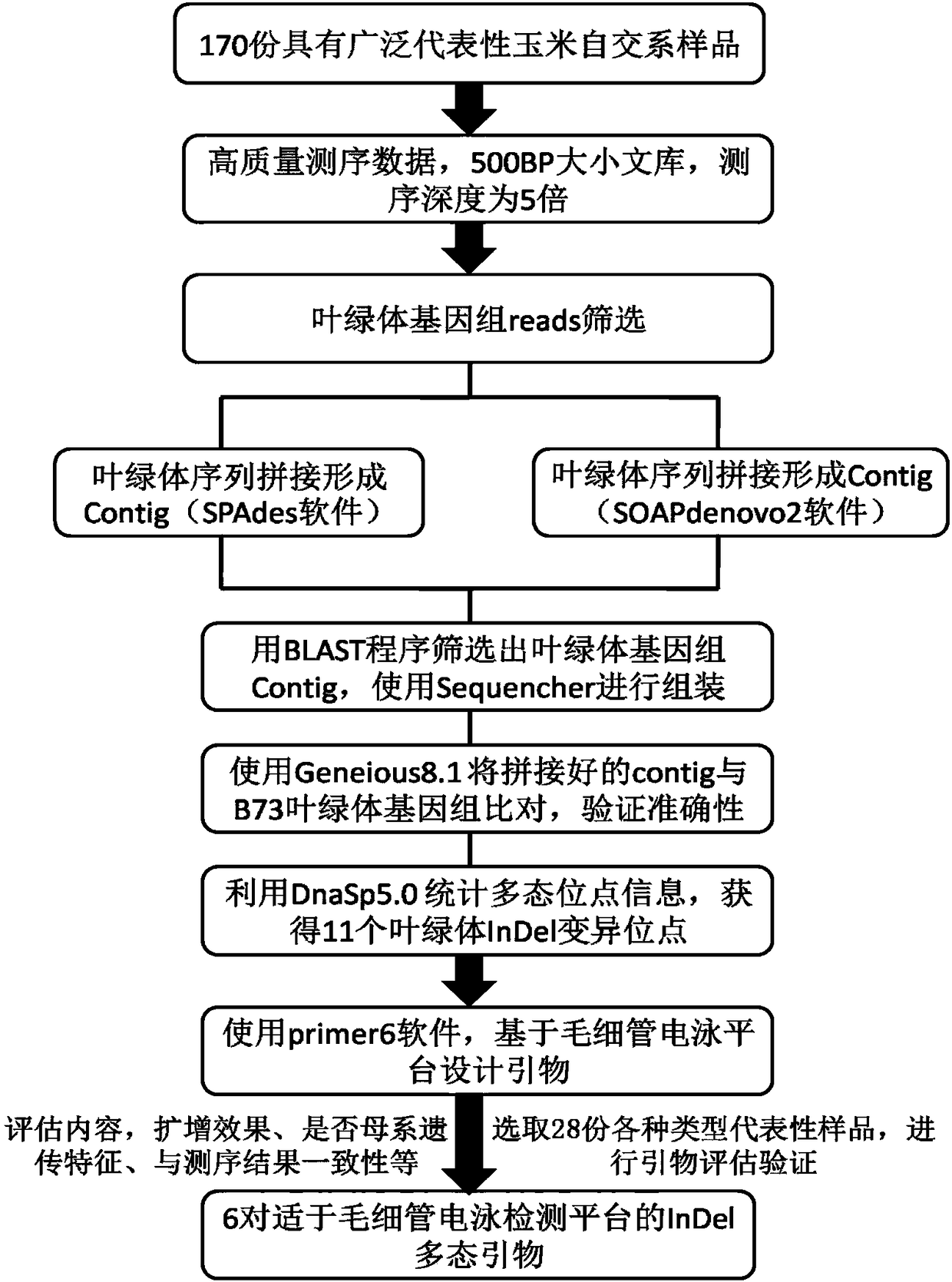
[0037] Example 1 Acquisition of InDel polymorphic sites and primers in maize chloroplast genome
[0038] Sample selection: 170 widely representative maize inbred lines were selected for whole genome sequencing. The 170 samples include common corn, waxy corn, sweet corn, popping corn and other corn types; including all heterotic groups in my country, including Tang Sipingtou, Luda Honggu, Ruide, Lanka, improved Ruide, improved Lanka, P group and local varieties.
[0039] Sample preparation: 170 maize samples were seeded in an incubator for 5 days, the first 3 days were no light conditions, and the last 2 days were light conditions (that is, sufficient light was given after unearthed). For each sample, leaves were selected from 30 green seedlings, mixed, and thoroughly ground under liquid nitrogen. Total DNA was extracted by CTAB method, and RNA was removed. The quality of the extracted DNA was detected by UV spectrophotometer and agarose electrophoresis, respectively. Agaros...
Embodiment 2
[0047] Example 2 Construction of maize variety chloroplast DNA-InDel fingerprint database using InDel polymorphic primers provided by the present invention
[0048] DNA extraction of maize varieties: each sample was seeded with seedlings, given light to form green seedlings. DNA extraction adopts the method of extracting DNA from mixed plants, mixing the green leaves of 30 individual plants. The specific steps of DNA extraction are carried out according to the standard of maize DNA molecular identification (Wang Fengge et al., 2014). Dilute the DNA to form a working solution with a concentration of 20ng / μL.
[0049] Synthesis of primers: The 6 pairs of primer sequences that were successfully verified according to Table 2, that is, the nucleotide sequences of the 6 pairs of primers determined in Example 1 are SEQ ID NO.13-14, SEQ ID NO.15-16, SEQ ID NO. 17-18, SEQ ID NO.19-20, SEQ ID NO.21-22, SEQ ID NO.23-24 were synthesized primers; the 5' end of one of each pair of primers ...
Embodiment 3
[0052] Embodiment 3 utilizes the maize chloroplast genome InDel polymorphic primer provided by the present invention to carry out maternal traceability analysis
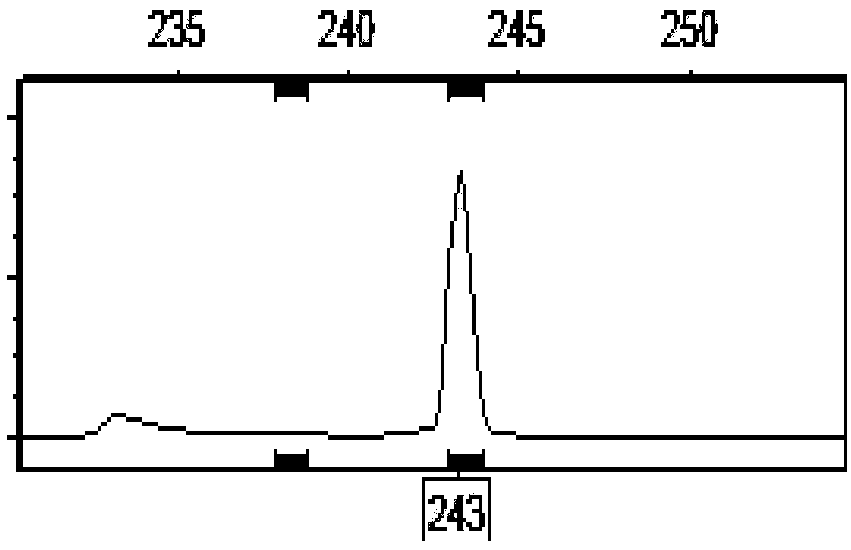
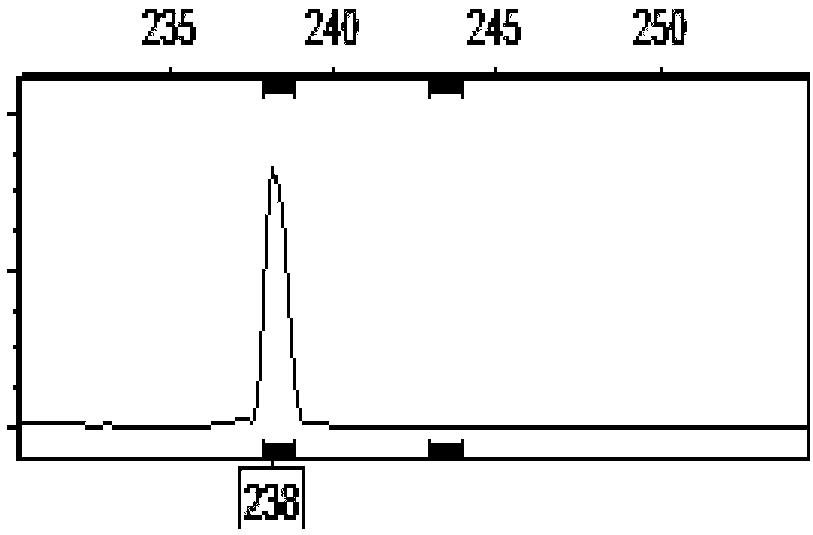
[0053] To be identified corn hybrid A, DNA was extracted from the green leaves of the seedlings, and the 6 pairs of InDel polymorphic primers confirmed in Example 1 of the present invention were used to carry out PCR amplification and fluorescent capillary electrophoresis to obtain fingerprint data. The specific method is the same as that in Example 2. Based on the comparison of the InDel fingerprint data of the A sample (Jingke 968) with the chloroplast InDel marker fingerprint database of the known maize inbred line varieties, it is determined that the chloroplast fingerprint information of the sample to be tested is the same as that of the B inbred line (Jing 724). (Jingke 968) is the parent of B (Jing 724).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com