Double-stranded RNA fragmentation method and use thereof
A technology of fragmentation and RNA virus, applied in the field of fragmentation and utilization of double-stranded RNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0139] The techniques disclosed below represent one of the aspects of the present invention for the purpose of being supported by experiments, which can be understood by those skilled in the art. The protection scope of the present application shall be interpreted according to the claims, and shall not be limited to the manner described in the item of the embodiment.
[0140] [Materials and methods]
[0141]
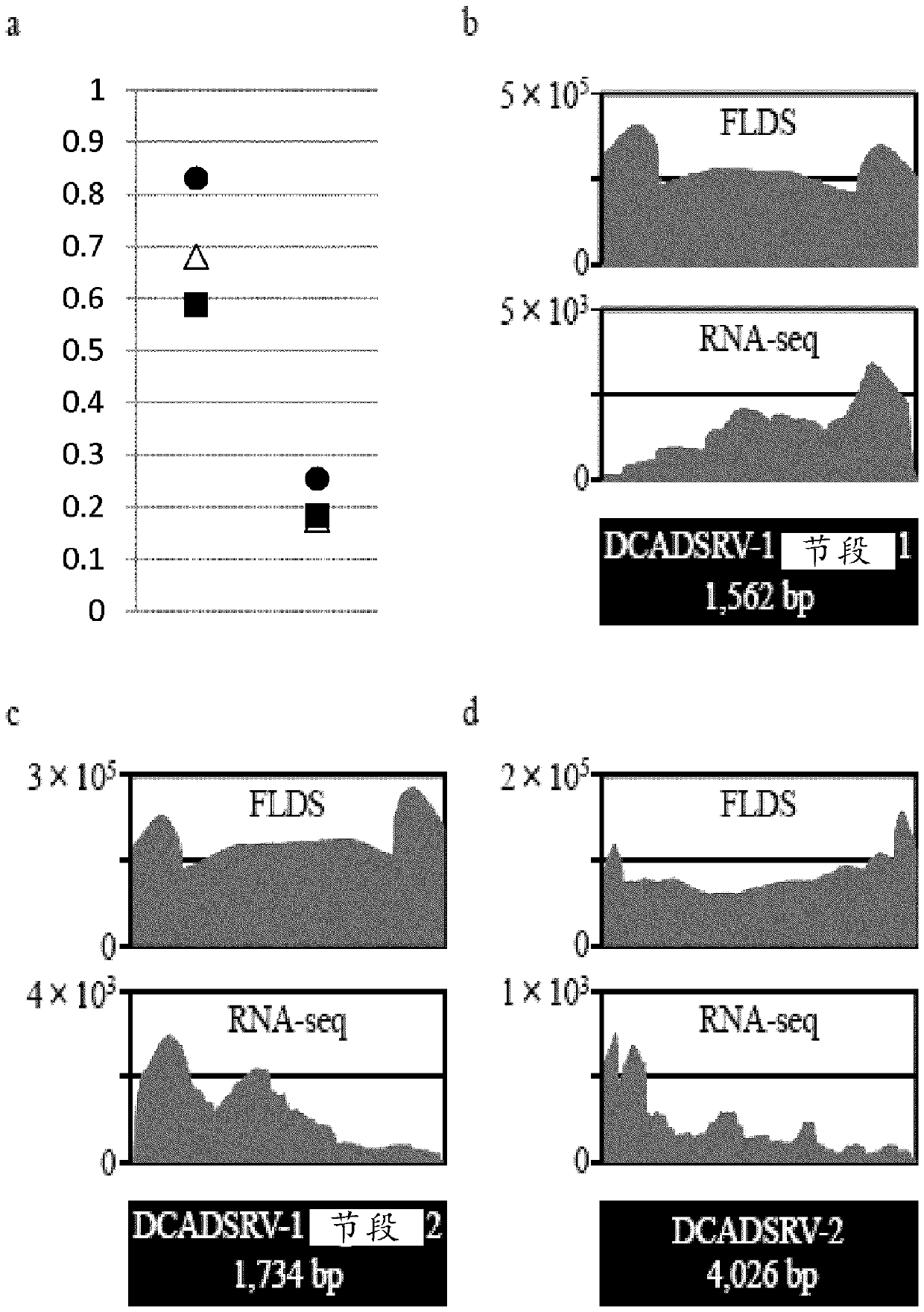
[0142] The rice blast fungus S-0412-II 1a infected with Magnaporthe oryzae chrysovirus 1 strain A (MoCV1-A) was planted in YG liquid medium (0.5% yeast extract, 2% glucose), at 25°C, 60r .p.m was cultured for 2 weeks with back-and-forth shaking. (this bacterium is to operate with the rice blast fungus produced in Vietnam, because it needs permission, so it is carried out under Professor Teraoka of Tokyo University of Agriculture and Technology with permission. In addition, about MoCV1-A, there are related patents or patent applications (such as , EP2679675, US2011-00...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com