DNA library construction method for fetus chromosome aneuploid detection
A technology for aneuploidy and library construction, which is applied in the field of non-invasive prenatal screening and detection, can solve the problems of high cost of library construction methods, poor practical operation results, and difficult replication operations, etc., so as to reduce the sequencing process and improve labor efficiency , time-saving effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] A DNA library construction method in the detection of fetal chromosomal aneuploidy, characterized in that, comprising:
[0040] Step 1: Physically interrupt the sample DNA to obtain a fragmented DNA solution;
[0041] Step 2: performing end repair and magnetic bead purification on the fragmented DNA to obtain purified blunt-ended DNA;
[0042] Step 3: performing adapter ligation on the blunt-ended DNA to obtain adapter-ligated DNA;
[0043] Step 4: performing PCR amplification on the DNA connected with the adapter to obtain a library;
[0044]Wherein, the step three specifically includes: sequentially adding blunt-end DNA, nuclease-free water, ligation buffer, dNTP, DNA ligase, P1 linker and tag sequence X into the centrifuge tube and mixing, and centrifuging and purifying after reaction.
[0045] The third step specifically includes:
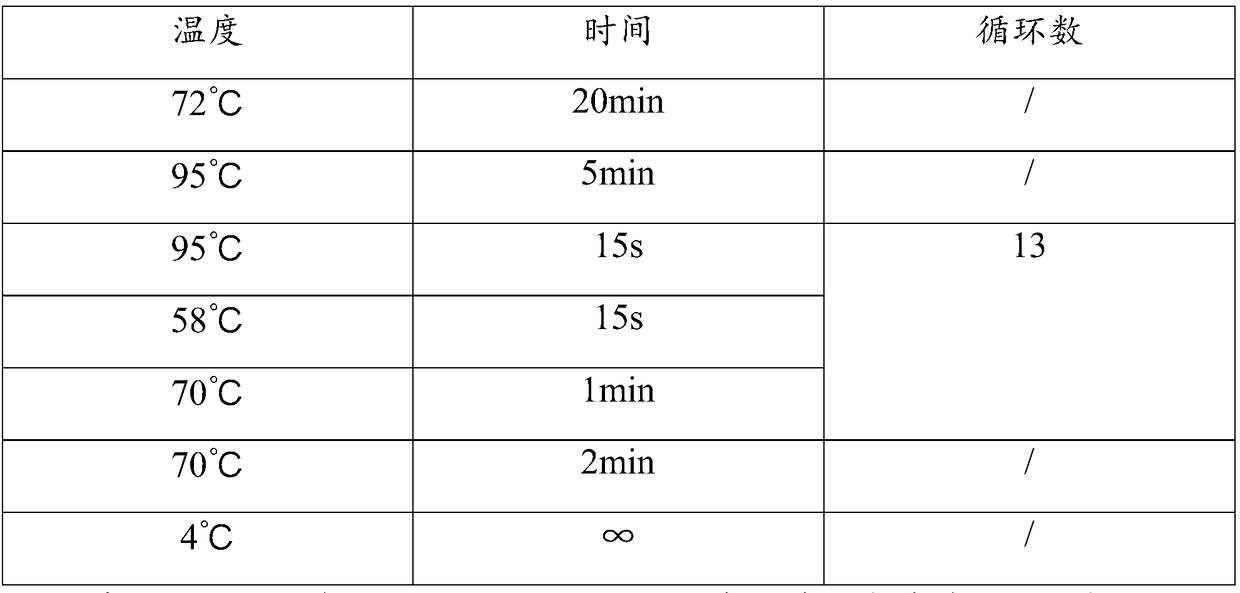
[0046] Step 1: Add 25ul blunt-end DNA, 16ul nuclease-free water, 5ul ligation buffer, 1ul dNTP, 1ul DNA ligase, 1ul P1 linker and 1u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com