Loop type enrichment detection primer of low-abundance mutation DNA and application thereof
A detection primer, low abundance technology, applied in recombinant DNA technology, DNA/RNA fragment, determination/inspection of microorganisms, etc., can solve the problems of low sensitivity, low content of mutated nucleic acid, high detection cost, etc. Highly specific and accurate effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Example 1: Enrichment detection primers for detection of T790M mutation and L858R mutation in the present invention
[0071] Table 1
[0072]
[0073]
[0074] In Table 1, T790M-Loop-F and L858R-Loop-F are the enrichment detection primers for T790M mutation and L858R mutation, where the underline is the base of the mutation site; T790M-R and L858R-R are the corresponding Downstream primers, the sequences of which are shown in SEQ ID NO:5-6.
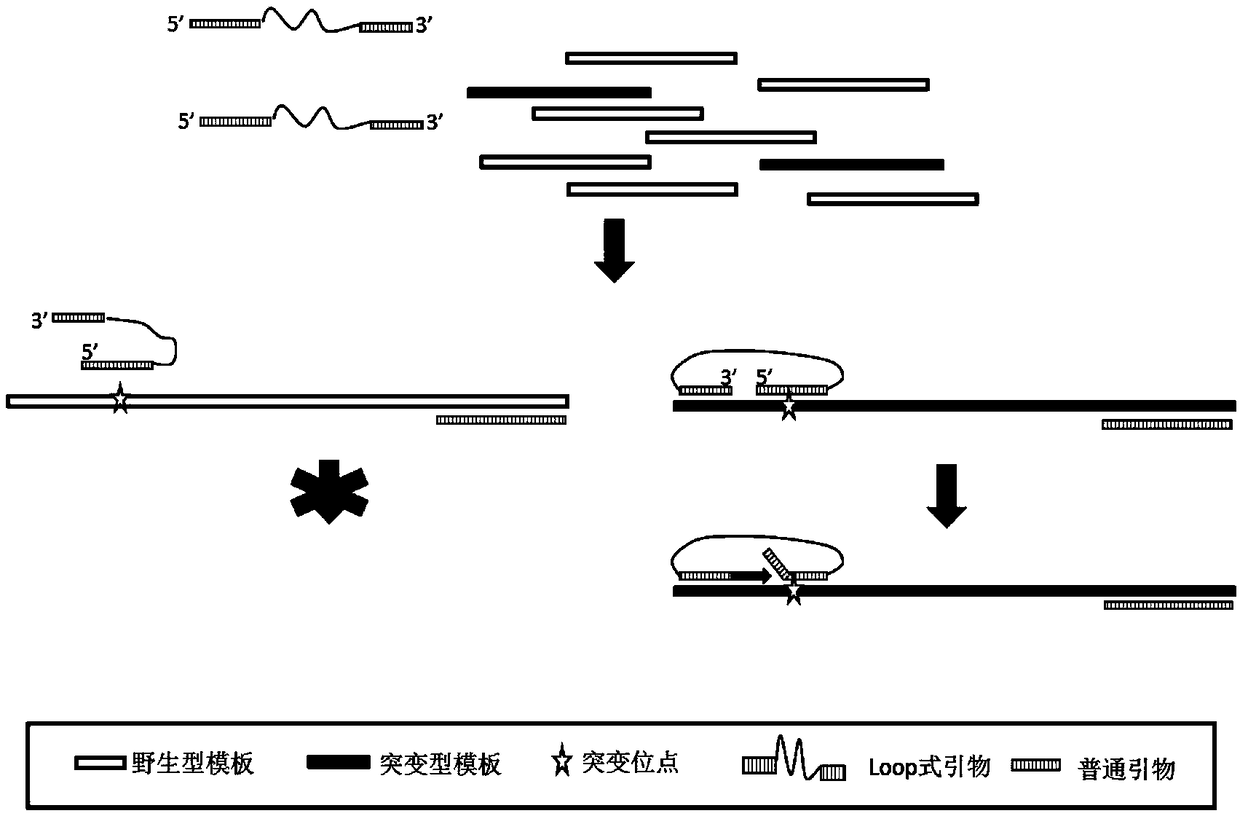
[0075] In T790M-Loop-F, the degree of polymerization of Oligo (dT) is 32, C12 means C12Spacer modification, TCATCATGCAGCTCA is the auxiliary primer nucleotide sequence (shown in SEQ ID NO: 1), and CACCGTGCAGC is the amplification primer nucleotide sequence (SEQ ID NO: 2), and the 3' end of the amplification primer and the 5' end of the auxiliary primer are separated from the template binding site by 0 bases. For the schematic diagram, see figure 2 .
[0076]In L858R-Loop-F, the degree of polymerization of Oligo (dT) is 28...
Embodiment 2
[0077] Embodiment 2: The present invention detects the enrichment detection kit (common PCR) of T790M mutation and L858R mutation
[0078] Table 2
[0079]
[0080]
[0081] In Table 2, CX-T790M-F and CX-T790M-R are T790M sequencing upstream and downstream amplification primers (shown in SEQ ID NO: 17-18), CX-L858R-F and CX-L858R-R are L858R sequencing The downstream amplification primers (shown in SEQ ID NO: 19-20), CX-F and CX-R are upstream and downstream general primers (shown in SEQ ID NO: 21-22) for upper-machine sequencing.
[0082] For T790M and L858R Mutation Enrichment Detection Kit (General PCR) consists of the following components: T790M Mutation Enrichment Primer Mix, L858R Mutation Enrichment Primer Mix, T790M Sequencing Amplification Primer Mix, L858R Sequencing Amplification Primer Mixture, sequencing upstream universal primer CX-F, sequencing downstream universal primer CX-R, mutation enrichment amplification reagent, sequencing amplification reagent, p...
Embodiment 3
[0093] Embodiment 3: The common PCR method that the present invention detects T790M mutation and L858R mutation
[0094] 1. Detection method
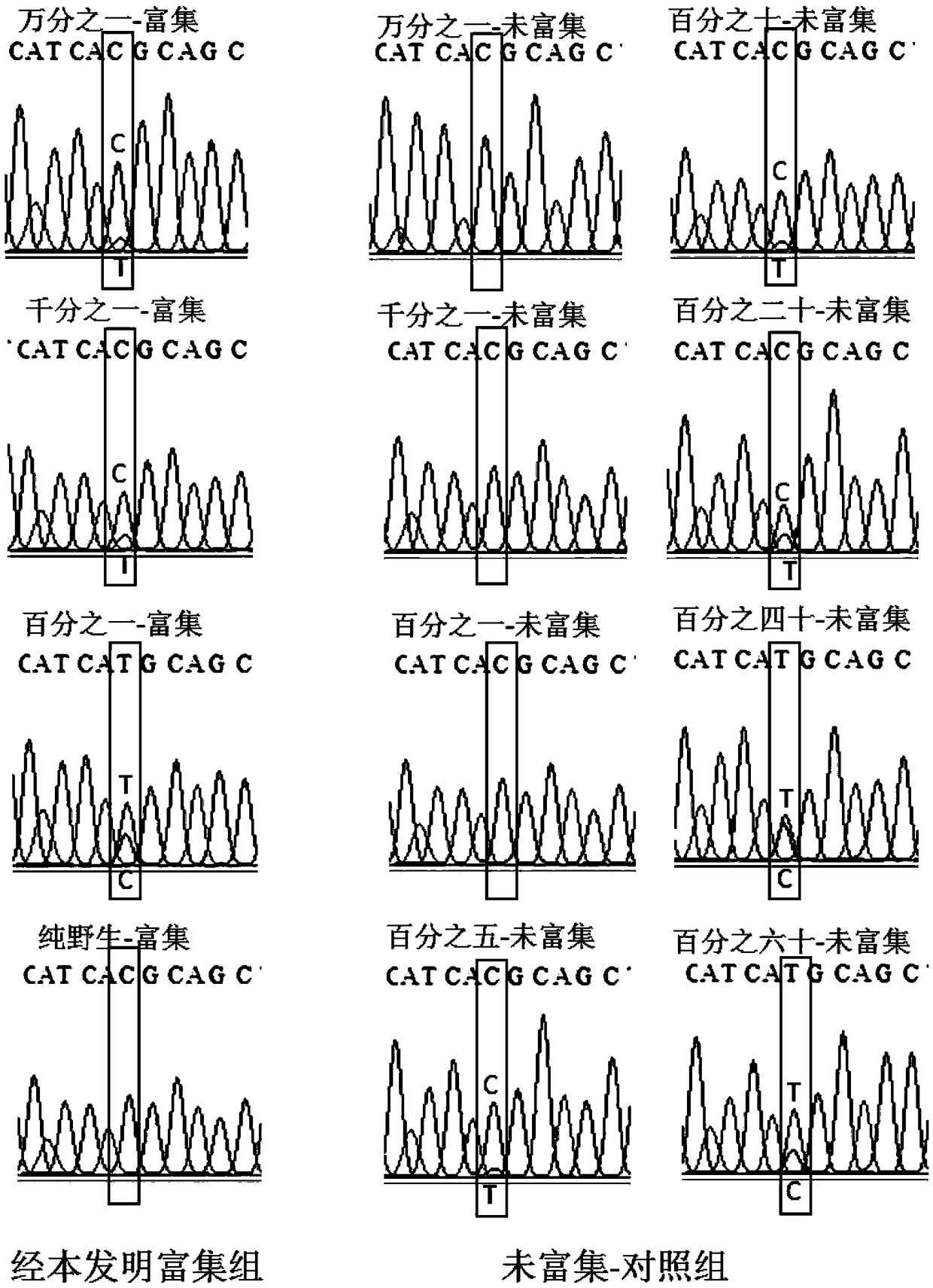
[0095] The kit in Example 2 was used to perform two rounds of amplification for detection. The first round of amplification was used to enrich low-abundance mutations, and the second round of amplification was used to add general sequencing primers to prepare for on-machine sequencing.
[0096] For the preparation of the first round of amplification system, take an eight-row PCR reaction tube, add 12.5 μl of the 2× detection system, 2.5 μl of the corresponding primer components, and 5-10 μl of the corresponding template, add water to 25 μl, mix well, Place the prepared PCR reaction tube in a PCR amplification instrument for amplification.
[0097] The first round of amplification procedure:
[0098] 95°C for 5min, cycle once
[0099] 95°C for 5s, 56°C for 10s, 68°C for 30s, cycle 15 times
[0100] 95°C 5s, 60°C 5s, 68°C 30s, cycle 3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com