Reagent for extracting total DNAs of Grifola frondosa and application thereof
A technology of Grifola frondosa and reagents, applied in the field of genus, can solve the problems of short DNA fragments, toxic reagents, and long time consumption, and achieve the effects of increased binding rate, high integrity, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
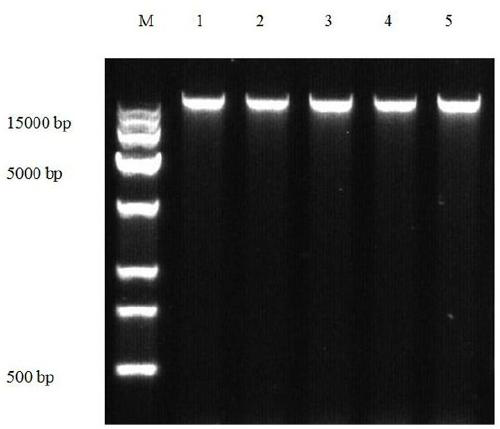
[0031] Embodiment 1, adopt this method to extract the total DNA of Grifola frondosa mycelium and fruiting body
[0032] Collect the mycelium and fruiting bodies of Grifola frondosa respectively, and the specific steps are as follows:
[0033] (1) Add 200 μL of BB to the DNA adsorption column, centrifuge at 10,000 rpm for 1 min, discard the waste liquid, and pretreat the DNA adsorption silica gel column;
[0034] (2) Grind 2-100 mg of the sample with dry ice, add 600 µL of LB, mix well, and centrifuge at 10,000 rpm for 30 s at room temperature;
[0035](3) Transfer the supernatant to a treated DNA adsorption column, centrifuge at 10,000 rpm for 1 min at room temperature, and discard the waste liquid;
[0036] (4) Add 600 µL of WB to the DNA adsorption silica gel column, centrifuge at 10,000 rpm at room temperature for 1 min, discard the waste liquid, put the DNA adsorption silica gel column back into the waste liquid collection tube, and centrifuge at 10,000 rpm at room temper...
Embodiment 2
[0046] Collect the total DNA of Grifola frondosa fruiting bodies, mycelium, and the total DNA of mushrooms, oyster mushrooms, and Cordyceps militaris respectively. The specific steps are as follows:
[0047] (1) Add 200 μL of BB to the DNA adsorption column, centrifuge at 10,000 rpm for 1 min, discard the waste liquid, and pretreat the DNA adsorption silica gel column;
[0048] (2) Grind 2-100 mg of the sample with dry ice, add 600 µL of LB, mix well, and centrifuge at 10,000 rpm for 30 s at room temperature;
[0049] (3) Transfer the supernatant to a treated DNA adsorption column, centrifuge at 10,000 rpm for 1 min at room temperature, and discard the waste liquid;
[0050] (4) Add 600 µL of WB to the DNA adsorption silica gel column, centrifuge at 10,000 rpm at room temperature for 1 min, discard the waste liquid, put the DNA adsorption silica gel column back into the waste liquid collection tube, and centrifuge at 10,000 rpm at room temperature for 1 min;
[0051] (5) Move...
Embodiment 3
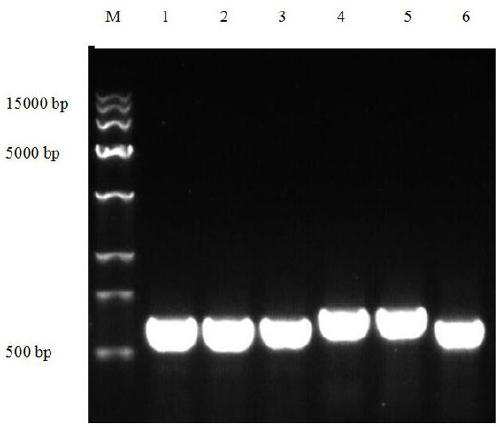
[0059] Embodiment 3, Grifola frondosa ITS sequence PCR amplification
[0060] ITS-4 / 5, a common primer for ITS fragments of edible fungi, was used to detect the genomes of Grifola frondosa fruiting body, mycelium, dry sample DNA and total DNA of shiitake mushroom, ganoderma lucidum and Flammulina velutipes.
[0061] The primer sequences are:
[0062] ITS-4: 5’-TCCTCCGCTTATTGATATGC-3’
[0063] ITS-5: 5’- GGAAGTAAAAGTCGTAACAAGG-3’
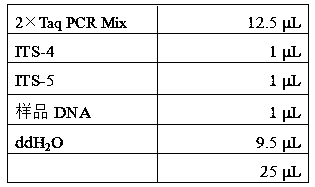
[0064] The reagents used for PCR amplification were purchased from Beijing Quanshijin Biotechnology Co., Ltd., and the reaction system was:
[0065]
[0066] The conditions of the PCR reaction are: 94°C 7min → [94°C 30s → 56°C 30s → 72°C 30s] ×34cicles → 72°C 10min → 4°C.
[0067] Agarose gel electrophoresis analysis
[0068] The total DNA and PCR amplification products were detected by 1% agarose gel electrophoresis, the loading buffer was 10×LoadingBuffer, the electrophoresis buffer was 1×TAE, the molecular weight standard marker was Trans 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com