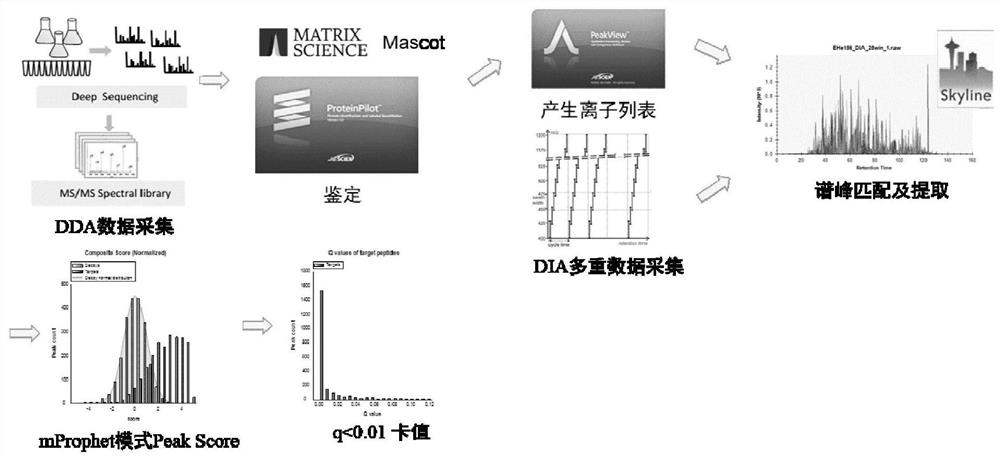
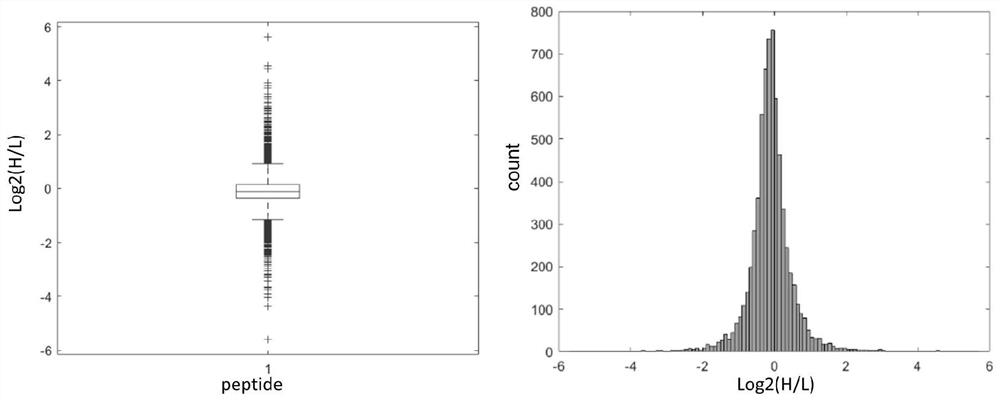
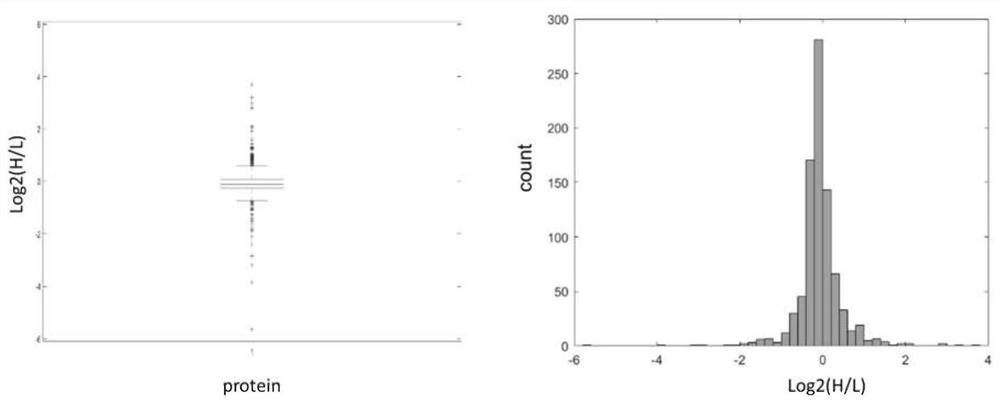
A method for protein quantification based on the dimethylation-tagged DIA strategy
A dimethylation and protein technology, which can be used in measurement devices, instruments, scientific instruments, etc., can solve the problems of low throughput and affect the quantitative accuracy, and achieve good reproducibility, improved quantitative accuracy, and high efficiency. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] 1. Protein extraction and sample pretreatment
[0030] E. coli samples were washed three times with 1×PBS. 8M urea + 0.1% protease inhibitor (cocktail, Sigma-Aldrich, MO) was used as the lysate to suspend the cell line. The cell lines were sonicated for 100 s with a tissue disruptor at a speed of 10,000. The cell extract was centrifuged at 25,000 g to obtain the supernatant, precipitated with acetone, discarded the supernatant, and evaporated to dryness. After reconstitution with 8M urea, the protein concentration was determined by Bradford method. Store at -20°C until use.
[0031] 1 mg E.coli protein extract was denatured and reduced with 2 μL 1M DTT at 56°C for 1 hour, then added 4 μL 1M IAA, and reacted in the dark for 40 minutes. Urea was then diluted to 1 M with 50 mM phosphate buffer (pH 8.0). Add trypsin to Ecoli according to 1:50 (enzyme / protein, mass ratio), and digest at 37°C overnight. Use a C18 trapping column to desalt and evaporate to dryness for la...
Embodiment 2
[0045] This strategy is suitable for absolute quantitative analysis based on internal standards. One labeling method is used to label the internal standard of the target protein, and the other labeling method is used to label the sample to be tested. The other processes are the same as in Example 1. The absolute value of the target protein of the sample to be tested is obtained according to the relative quantification results between the two markers of the target protein, and then the absolute quantification of the target protein based on the DIA strategy is realized. This strategy achieves high-accuracy absolute quantification of target proteins.
Embodiment 3
[0047] High-throughput DIA quantification can be achieved by dimethylated pentagrams. The five samples to be tested for relative quantitative comparison were respectively marked with dimethylation, and the marking method was: sample1:CH 2 O+NaBH 3 CN(Dim28); sample2: CH 2 O+NaBD 3 CN(Dim30); sample3: 13 CH 2 O+NaBD 3 CN(Dim32); sample4: 13 cd 2 O+NaBH 3 CN(Dim34); sample5: 13 cd 2 O+NaBD 3 CN(Dim36). DIA data collection is carried out after mixing, and other processes are the same as in Example 1, and the parent-child ion pairs (Dim28, Dim30, Dim32, Dim34, Dim36) of the fivefold label are respectively extracted for comparison, that is to say, simultaneous DIA of 5 samples in one experiment Quantitative analysis has obtained high-accuracy quantitative information of large-scale proteomes.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com