Screening method for protein nucleic acid aptamer
A nucleic acid aptamer and screening method technology, which is applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of low aptamer enrichment efficiency, aptamer affinity and specificity reduction, and achieve Good enrichment, high affinity, and improved screening efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
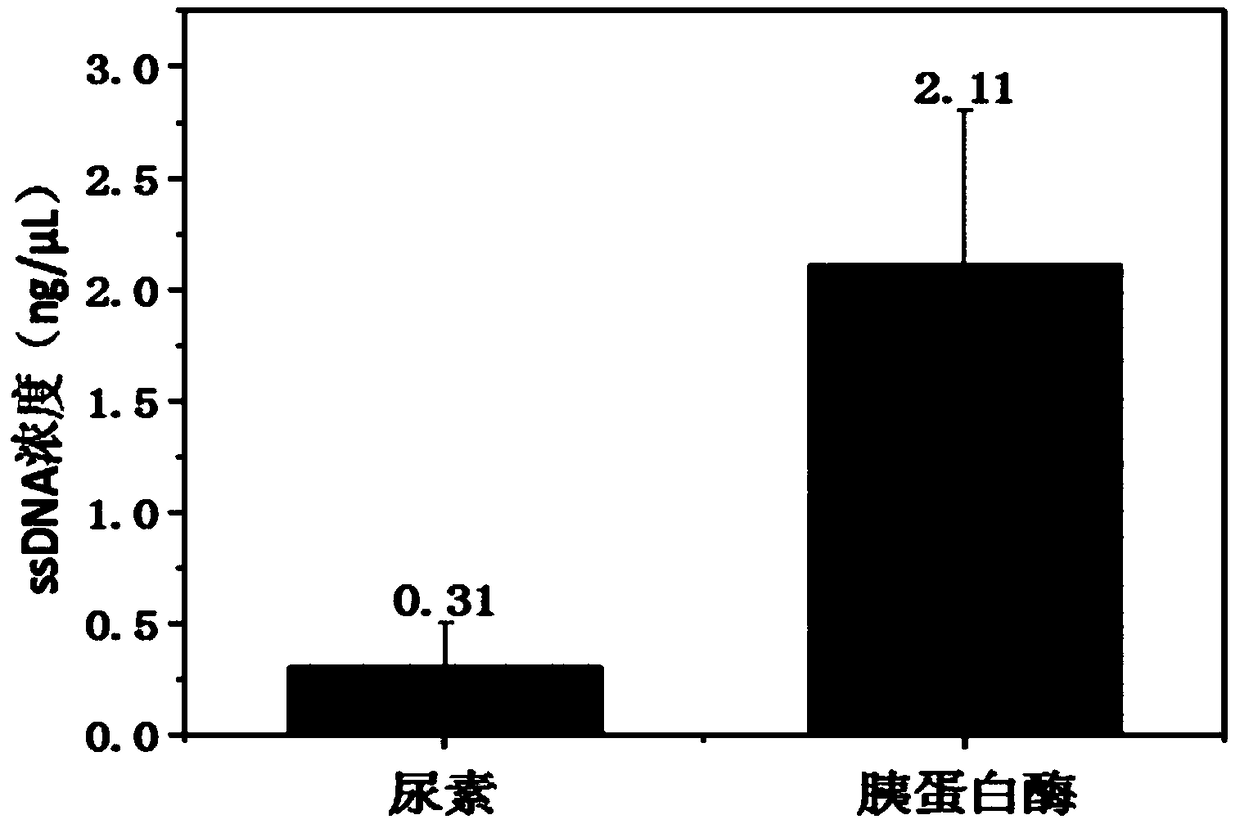
[0035] Example 1: A screening method for protein nucleic acid aptamers: embed the protein on the surface of a polystyrene 96-well plate or other utensils that absorb protein; add trypsin or protease to digest and degrade the protein, and make the protein and oligonucleotide The acid-binding complex is detached from the surface of the container, the protease is inactivated by heating and the ssDNA is released; the ssDNA that specifically binds to the protein is enriched, and aptamers with high affinity are screened.
Embodiment 2
[0036]Example 2: A screening method for protein nucleic acid aptamers: the specific steps are as follows: protein molecules are coated in a 96-well polystyrene plate, blocked with BSA or FBS, and then the oligonucleotide sequence ssDNA is added to the Incubate in the plate wells coated with protein for a certain period of time, remove the unbound oligonucleotide sequences with buffer, digest the protein-oligonucleotide complexes in the 96-well polystyrene plate with trypsin diluent, and then Boil the entire solution system in boiling water until the protein is denatured; centrifuge at 12,000g for 20 minutes to remove the precipitate to remove protein residues; use PCR to enrich the secondary library, use asymmetric PCR and denaturing polyacrylamide gel cutting to prepare and purify the oligo Nucleotide single strand; enriched nucleic acid aptamers can be obtained after cycling the above steps 7-10 times.
Embodiment 3
[0037] Embodiment 3, the screening method experiment of protein nucleic acid aptamer:
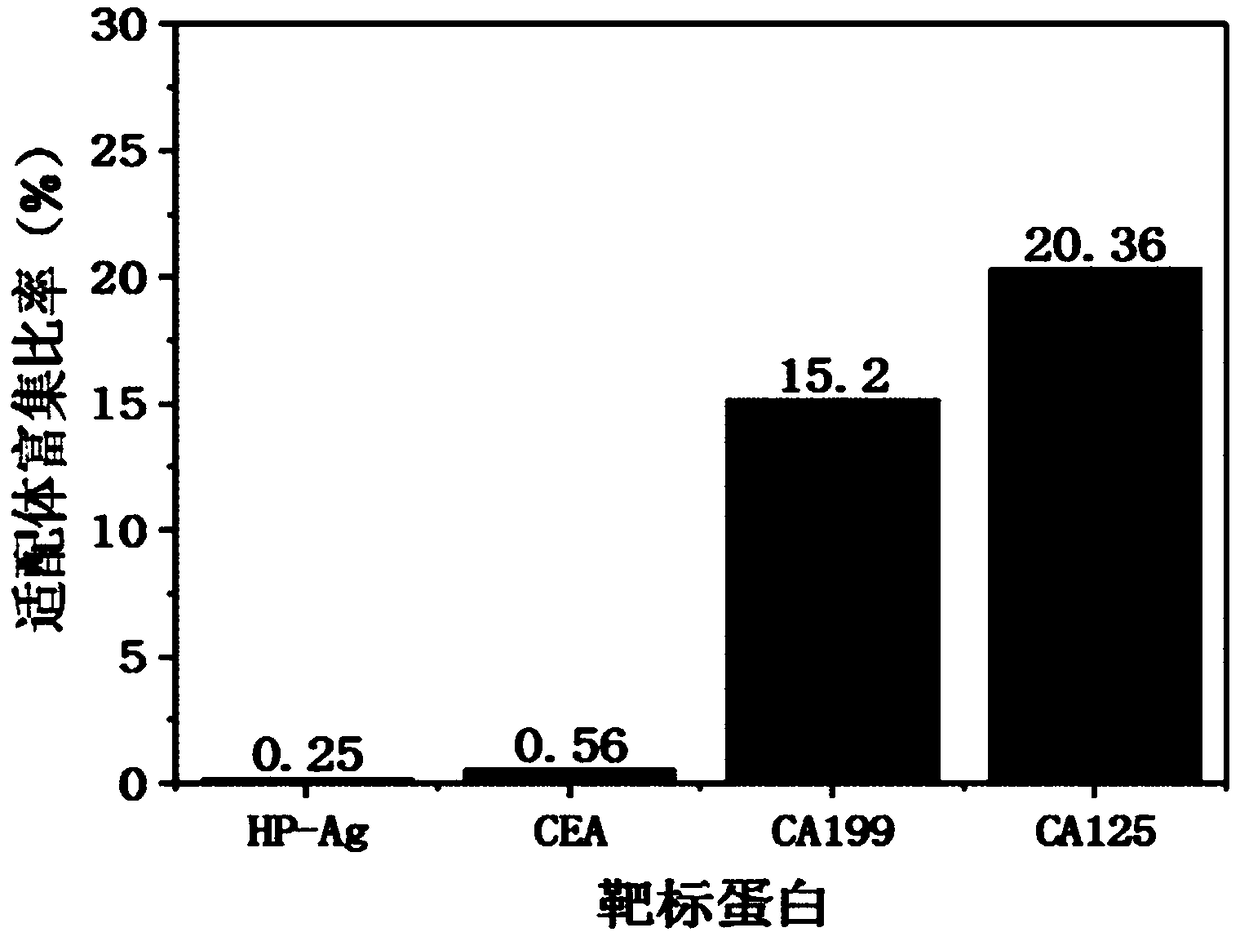
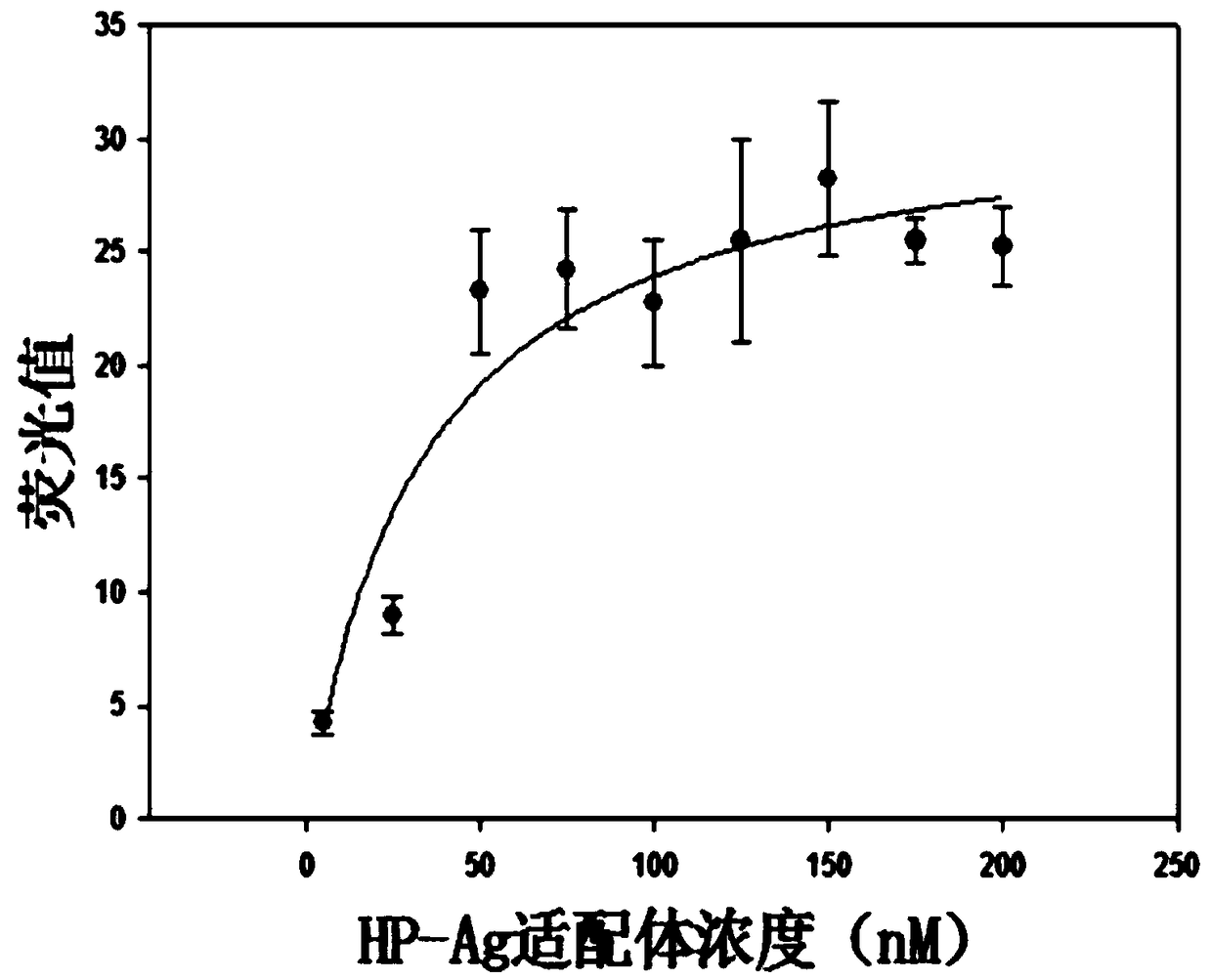
[0038] In this example, the surface recombinant antigen HP-Ag of Helicobacter pylori, natural carcinoembryonic antigen CEA, and tumor markers CA125 and CA199 were used as targets, and the following screening methods were used to screen nucleic acid aptamers. The specific steps are as follows:
[0039] 1 Synthesize 80nt random single-stranded DNA library and primers;
[0040] Random library: 5'-AAC TAC GCT CAG ACA GAC AC-40N-CTG TGA TGC GAC TAT CTG AG -3';
[0041] 5' Primer: 5'-AACTACGCTCAGACAGACAC-3'
[0042] 3' Primer: 5'-CTCAGATAGTCGCATCACAG-3'
[0043] 2 Screening: Dilute the four proteins with protein coating solution and coat them in 96-well polystyrene plates, shake slowly at 37°C for 2 hours, remove the solution and wash with PBS, add an equal volume of 1mg / mL bovine serum albumin, After shaking slowly at 37°C for 1 h, the protein solution was discarded and the 96-well polystyren...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com