Cronobacter sakazakii CRISPR typing method
A typing method and bacillus technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problem of no suitable CRISPR typing method for Cronobacter sakazakii, etc., to facilitate the molecular traceability and operation of strains Simple and convenient, low equipment requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
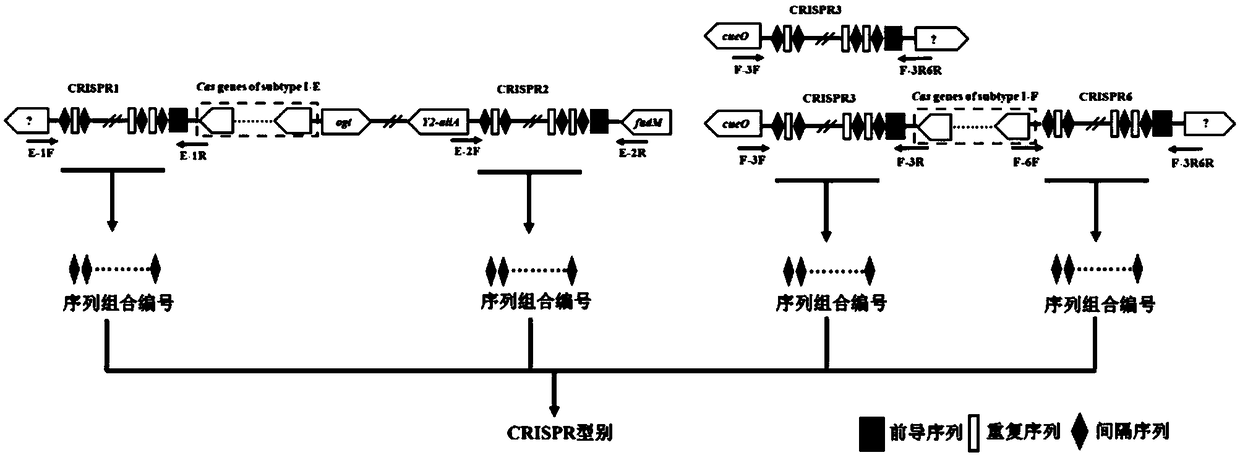
[0034] Example 1: CRISPR typing method of 136 strains of Kronobacter sakazaki, and its resolution comparison with MLST typing method
[0035] (1) Strain
[0036] The strain used in this experiment was selected from 136 strains of Kronobacter sakazaki isolated from various regions of our country preserved in our center. Traditional biochemical identification and molecular testing have been completed and it is determined to be Kronobacter sakazaki.
[0037] (2) Extraction of bacterial DNA
[0038] Magen’s bacterial genomic DNA extraction kit was used to extract the genomic DNA of the 136 strains to be tested. Kronobacter sakazaki ATCC 29544 was used as a positive control, ddH 2 O is a negative control. The extracted DNA is stored at -20°C.
[0039] (3) Primer synthesis
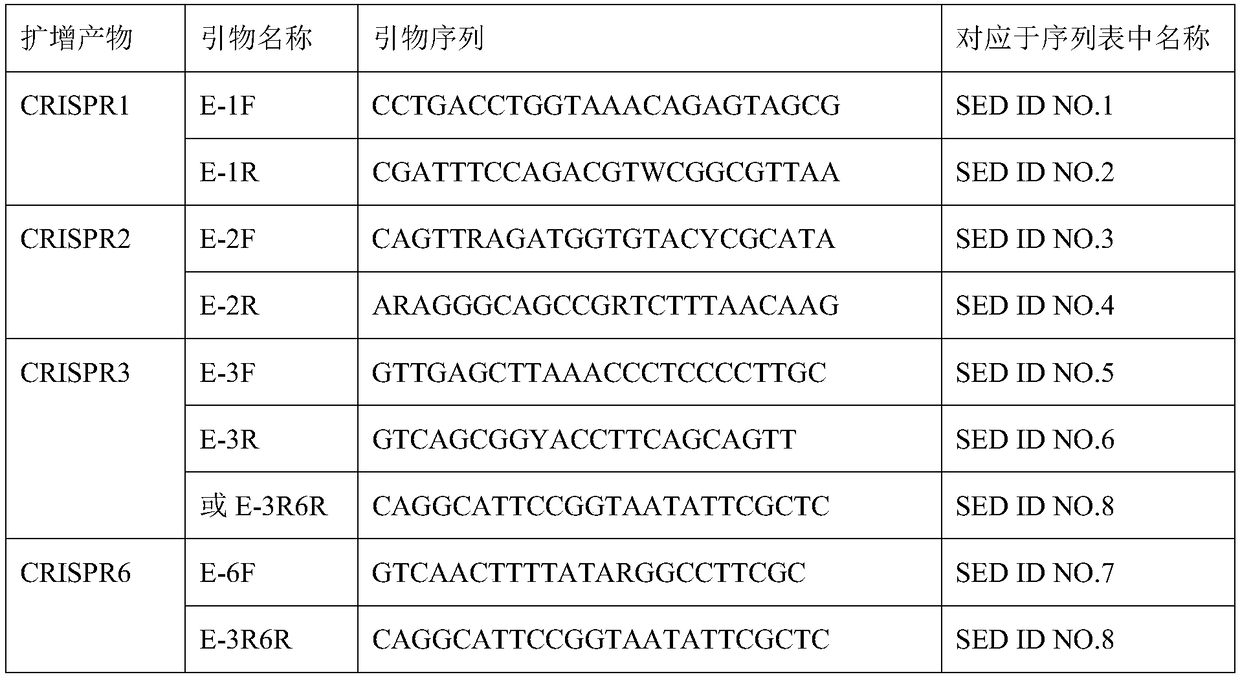
[0040] Table 1 Primer list for CRISPR molecule amplification
[0041]
[0042] The primer concentrations used in PCR amplification are all 10 μmol / L.
[0043] (4) PCR amplification
[0044] 4.1 CRISPR1 amplification PCR reac...
Embodiment 2
[0079] Example 2: Comparison of molecular traceability of CRISPR typing and MLST typing methods
[0080] (1) Download the full-length genomes of 23 strains (including ST4, ST12 and ST13) isolated from patients and feeding formula after the outbreak of Cronobacter Sakazaki in the French neonatal intensive care unit in 1994 Sequence (Masoodet al. BMC Genomics 16:750), according to the method in Example 1, determine the type and number of CRISPR molecules, and extract the spacer sequence for typing.
[0081] (2) Three ST-type strains of ST4, ST12 and ST13 in Example 1 were selected for comparison of CRISPR typing and MLST typing methods, and the results are shown in Table 3 below.
[0082] Table 3 Comparison of CRISPR and MLST typing results of 38 Cronobacter sakazaki strains
[0083]
[0084]
[0085] As shown in Table 3, three ST-type strains isolated from patients in the French neonatal intensive care unit outbreak of Kronobacter Sakazaki infection are ST4 (13 strains), ST12 (5 strain...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com