Nucleic acid aptamer specifically recognizing vimentin and its application
A nucleic acid aptamer and specific technology, applied in the biological field, can solve the problems of high immunogenicity, high production cost, batch difference, etc., and achieve the effects of simple reaction conditions, short incubation time and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
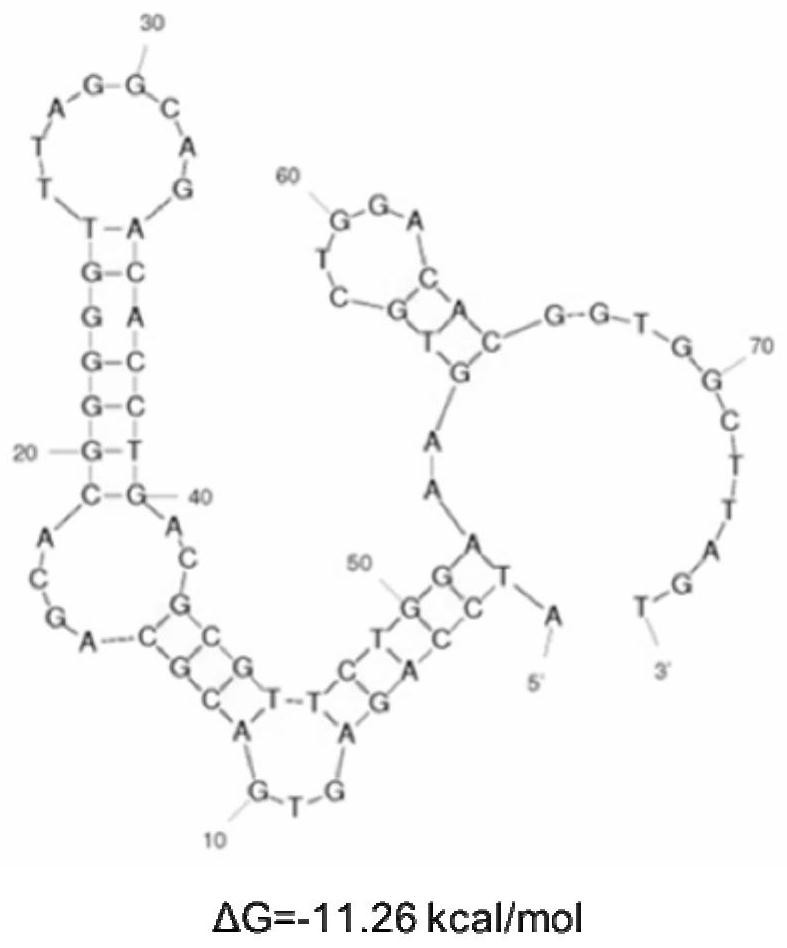
[0060] Example 1. Screening of nucleic acid aptamers
[0061] 1. Design and synthesis of random library
[0062] Design and synthesize a random library with 18 fixed nucleotides at both ends and 40 nucleotides in the middle: 5'- ATCCAGAGTGACGCAGCA -N(40)- TGGACACGGTGGCTTAGT -3'; N(40) represents a random nucleotide sequence of 40 A, T, C or G.
[0063] 2. SELEX screening
[0064] The SELEX screening process is divided into two parts: positive screening and negative screening. Specific steps are as follows:
[0065] 1. Positive screening
[0066] 10 μg of recombinant human Vimentin protein (6X histidine tag) was incubated with an appropriate amount of nickel agarose beads (Ni-SepharoseBeads) for 30 minutes at room temperature, and then coupled to nickel agarose beads, Binding BufferⅠ (1%BSA, 0.1%Tween) -20, 0.2 mg / mL tRNA, DPBS, pH 7.4) resuspended for use, washed to remove unbound substances, and used it as the target for forward screening. The 10 OD (about 14 nmol) r...
Embodiment 2
[0082] Example 2. Binding ability of nucleic acid aptamer AptVim to Vimentin protein
[0083] 1. Flow cytometry and Dot Blot experiments to detect the binding of nucleic acid aptamer AptVim to Vimentin protein
[0084] The 5'-end Cy3-modified AptVim sequence and the 5'-end biotin-modified AptVim sequence were synthesized according to the method in Example 1, respectively, and their binding to Vimentin protein was detected by flow assay and Dot blot assay. The specific detection method is the same as the method in Step 3 of Embodiment 1.
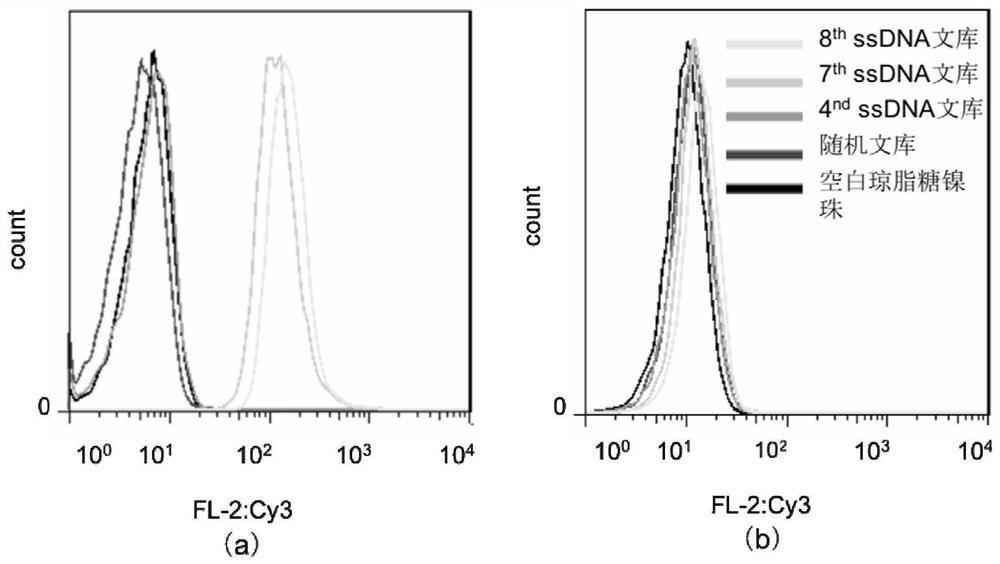
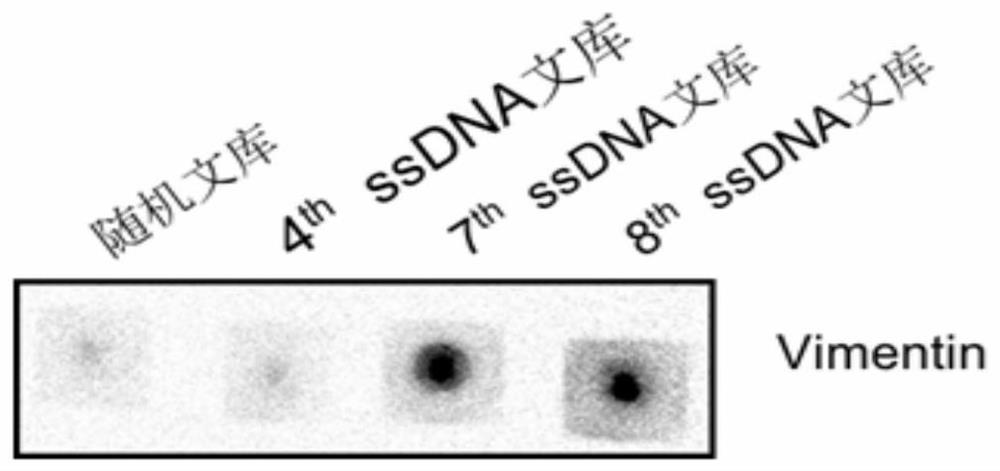
[0085] The flow detection results and Dot Blot detection results are respectively as follows Figure 4 and Figure 5 shown. The results showed that the nucleic acid aptamer AptVim was obviously bound to Vimentin protein, but not to 6X histidine polypeptide.
[0086] 2. Affinity determination of nucleic acid aptamer AptVim
[0087] In order to determine the affinity of the nucleic acid aptamer AptVim and Vimentin protein, the final workin...
Embodiment 3
[0099] Example 3. Aptamer AptVim induces apoptosis of Vimentin-positive cells
[0100] Panc-1 cells and BxPC-3 cells were seeded in 6-well plates for overnight culture, and divided into the following groups according to different treatment reagents:
[0101] AptVim+Turbofect treatment group: The nucleic acid aptamer AptVim (the concentration of nucleic acid aptamer AptVim was adjusted to 1 μM with serum-free DMEM) was coated with Turbofect, and the cells were treated for 24 hours;
[0102] Turbofect treatment group: Turbofect treated cells for 24 hours;
[0103] Turbofect+random library treatment group: After the random library (serum-free DMEM adjusted the random library concentration to 1 μM) was coated with Turbofect, the cells were treated for 24 hours;
[0104] AptVim treatment group: cells were directly treated with nucleic acid aptamer AptVim for 24 hours without Turbofect wrapping, and the final concentration of nucleic acid aptamer AptVim was 1 μM.
[0105] NC contr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com