Method for detecting hsa_circ_0007986 as novel biomarker in serum of patient with esophagus cancer and application
A technology of biomarkers and detection methods, applied in the field of tumor molecular biology, can solve the problems of unsatisfactory curative effect, loss of surgical indications, and lack of specific symptoms of esophageal cancer, achieving easy recurrence and metastasis, easy dynamic monitoring, high Effects of sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
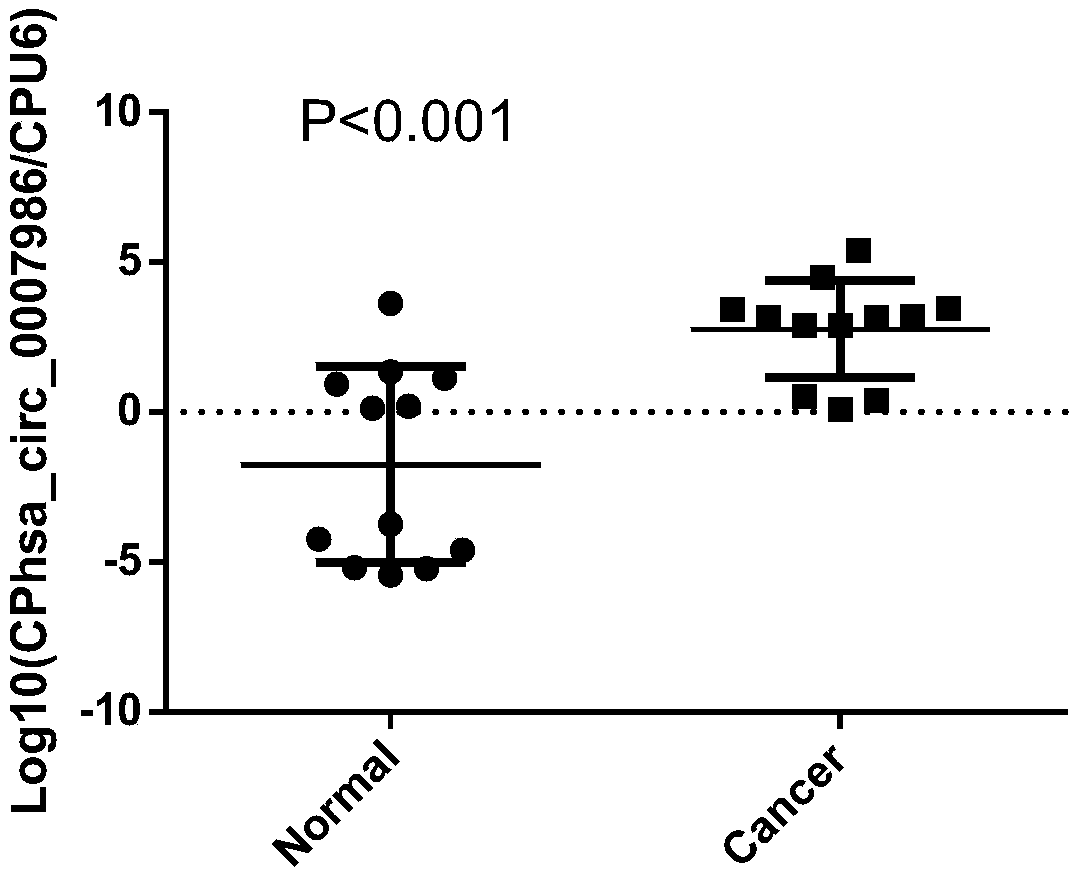
[0023] Example 1 Verification of the differential expression of hsa_circ_0007986 in esophageal squamous patients and healthy subjects
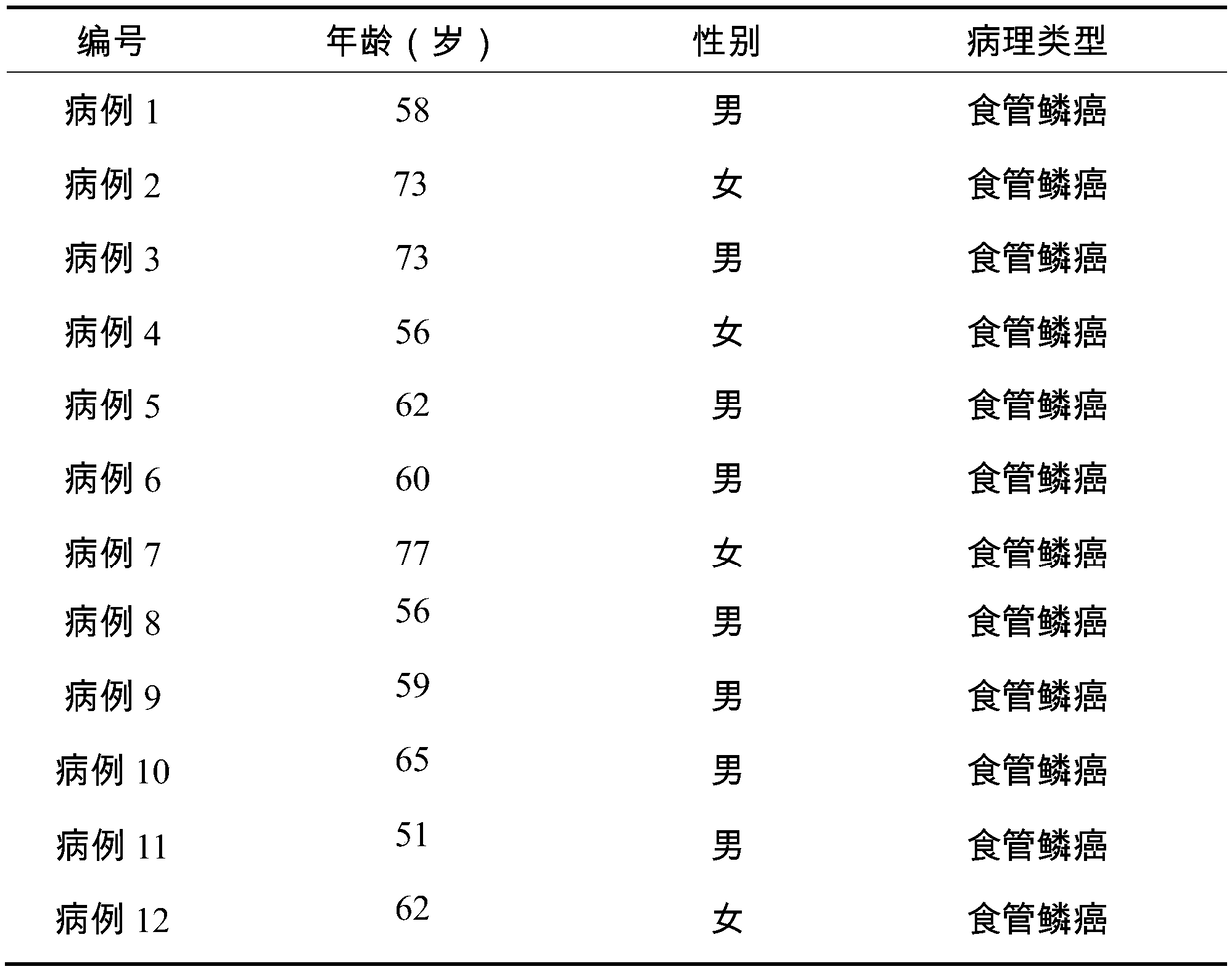
[0024] 1. Collection of serum samples: Each esophageal cancer patient and control group provided 5 mL of fresh blood, collected the upper serum after centrifugation, and stored at -80°C. The controls were all healthy subjects, and the experimental sample information is shown in Table 1 and Table 2.
[0025] Table 1: Basic information of 12 patients with esophageal cancer provided serum
[0026]
[0027] Table 2: Basic information of 12 normal controls
[0028]
[0029]
[0030] 2. For the extraction of serum RNA, refer to the operation procedure in the instruction manual of TRIzol LS Reagent for specific operation steps.
[0031] 3. Reverse transcription: reverse transcription of serum RNA into cDNA, the reverse transcription reaction system is as follows, see Table 3:
[0032] Table 3: Reverse transcription reaction system
[003...
Embodiment 2
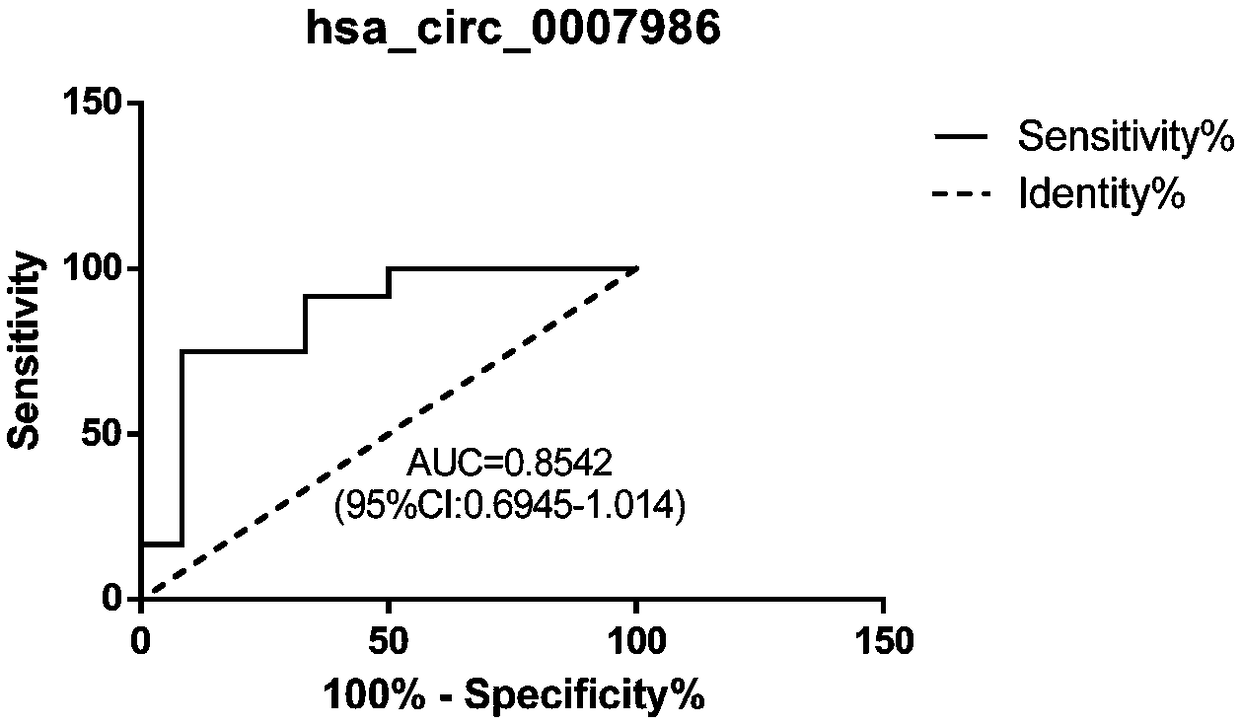
[0045] Example 2 Verification of hsa_circ_0007986 as a predictive reagent for esophageal cancer
[0046] 1. Composition of digital PCR kit
[0047] (1) Forward primer: such as SEQ ID NO:2; Reverse primer: such as SEQ ID NO:3;
[0048] (2) Internal reference U6 primer: the forward primer is shown in SEQ ID NO:4; the reverse primer is shown in SEQ ID NO:5;
[0049] (3) For other reagents, refer to SYBR Premix Ex TaqTM II (Tli RNaseH Plus) Quantitative Kit (Code No. RR820A).
[0050] 2. Perform digital PCR detection of hsa_circRNA-0007986 reagent
[0051] (1) Preparation of serum RNA
[0052] Serum samples from another 20 patients with esophageal squamous cell carcinoma were selected, and the specific operation steps of RNA extraction were referred to the operation procedure in the instruction manual of TRIzol LS Reagent. The purity and concentration of the extracted RNA were quantified by NanoDrop ND-1000 Nucleic Acid Quantifier (NanoDrop Technologies, Wilmington, Delaware),...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com