Composition for detection and detection method
A detection method and composition technology, applied in the field of molecular biology, can solve problems such as high cost, time-consuming and labor-intensive, and inability to meet the quantitative requirements of the transgenic labeling system
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] Example 1. Establishment of a novel fluorescent multiplex ligation-dependent probe amplification technology system F-MLPA
[0076] (1) Design of ligation probe sequence
[0077] According to the whole or partial sequence of the target nucleotide sequence to be detected or to be amplified (hereinafter referred to as sequence A), obtain a nucleotide sequence complementary to the target nucleotide sequence to be detected or to be amplified (hereinafter referred to as sequence C) ), the complementary binding nucleotide sequence is divided into two parts, that is, two nucleotide sequences (hereinafter referred to as sequence C1 and sequence C2, it should be pointed out that sequence C1 and sequence C2 have no sequence, but simply Used to represent the different parts in the two parts, or the different nucleotide sequences in the two nucleotide sequences), one part (sequence C1) is used to design the upstream connection probe, and the other part (sequence C2) Used to design ...
Embodiment 2
[0122] Embodiment 2, condition optimization experiment
[0123] (1) Optimization of the DNA denaturation process during the ligation probe reaction
[0124] The single-strand preparation process, that is, the DNA denaturation process in the kit manual of the Dutch MRC-Holland company is 95°C for 5 minutes, and then cooled to 25°C.
[0125] In Example 1, the DNA denaturation process was changed to 98°C for 5 minutes, and then quickly placed in ice water for 20 minutes.
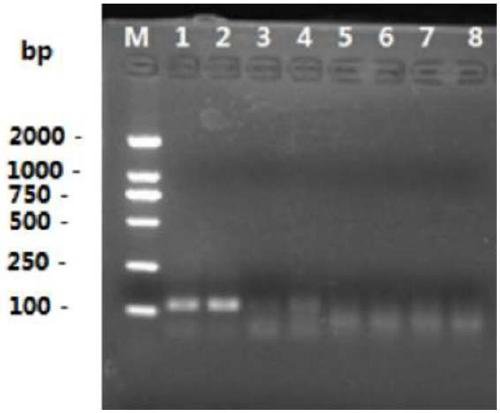
[0126] Using the transgenic crop MON810 as the template and MON810 as the target gene, the DNA denaturation process described above was compared and optimized. 2% agarose gel electrophoresis detection results.
[0127] DNA denaturation process optimization results such as figure 2 shown. From figure 2 It can be seen from the figure that if the DNA denaturation process is changed to 98°C for 5 minutes, and then quickly placed in ice water, the amplified product of the ligation product is clearer.
[0128...
Embodiment 3
[0134] Embodiment 3, single weight and multiple feasibility verification experiments
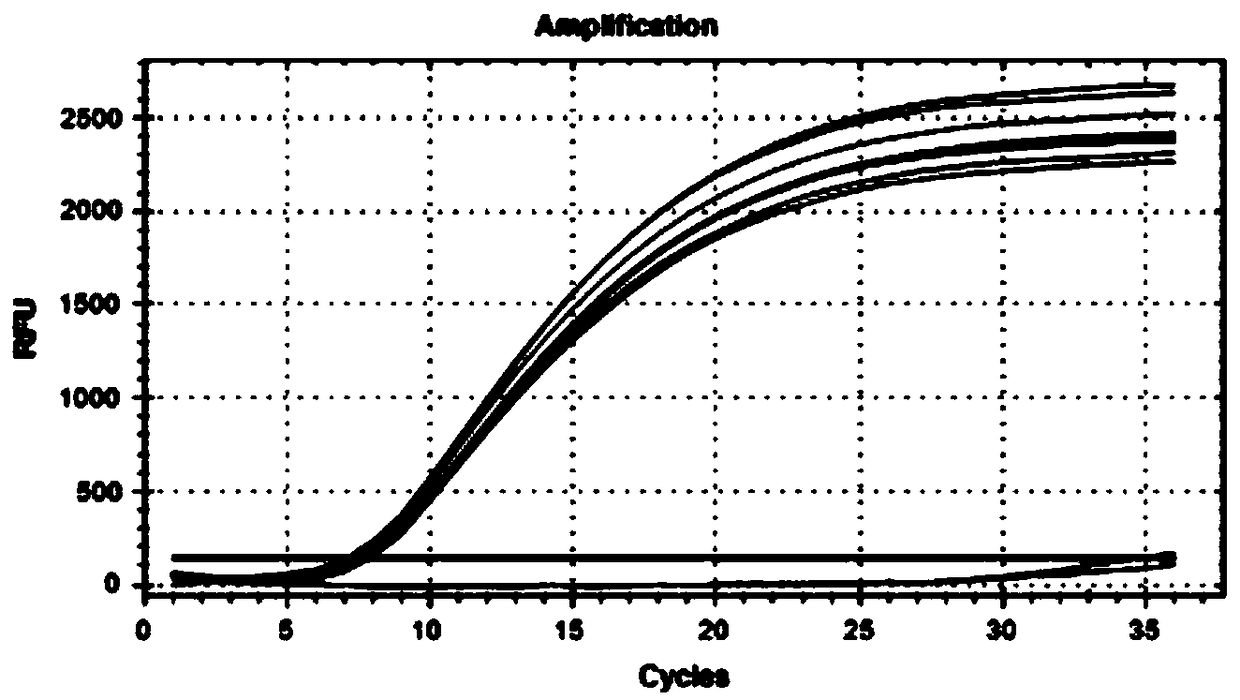
[0135] (1) Single feasibility verification experiment
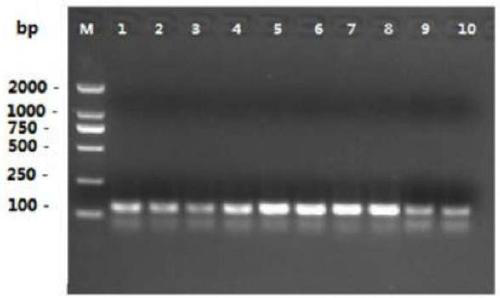
[0136] The feasibility of multiple pairs of connection-dependent probes shown in Table 2 in Example 1 was verified respectively. The template and connection probe sequences in each reaction system of the feasibility verification experiment were adjusted according to Example 1, and the others were consistent with Example 1. Finally, the experimental results were detected by 2% gel electrophoresis.
[0137] The results of the single-fold feasibility verification experiment are as follows: Figure 5 shown. From Figure 5 It can be seen that the four groups of probes (Mon810-2F and Mon810-2R, ZSS-2F and ZSS-2R, GTS-2F and GTS-2R, LEC-2F and LEC-2R) are all effectively connected and the connection products can be used in general PCR amplification was carried out under the guidance of primers, and the fragment sizes of the amplified produ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com