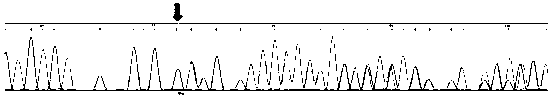
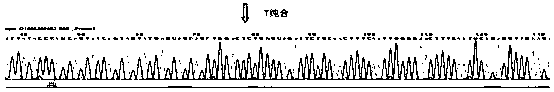
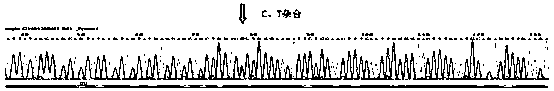
Primer, method and kit for detecting mutation sites of human BRCA1 and BRCA2 gene all-coding sequences
A mutation site and gene encoding technology, applied in the field of gene mutation detection in biotechnology, can solve the problems of low sensitivity, long cycle and high cost, and achieve the effects of improving primer specificity, simple operation and cost saving.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] The performance verification of this kit is carried out by detecting the standard of known mutation type and frequency.
[0037] A. Standard DNA Quantification
[0038] Use Novagen human genomic DNA (Merck, Cat. No. 69237) as a standard DNA template, use Fluorescence Quantitative Instrument and Corresponding The quantification reagent (Invitrogen) was used for quantification, and the concentration was 100 ng / uL.
[0039] B. The multiplex PCR method amplifies the target region of the sample genome.
[0040]1). Prepare a sterile, nuclease-free 200 μL PCR tube and place it on ice; prepare the PCR reaction system as shown in the table below, and be careful to avoid cross-contamination.
[0041] components
volume
2×PCR mix
6.25 μL
specific primer
2.0 μL
Universal Primer R5
0.5μL
Universal Primer R7
0.5μL
DNA template, 50ng
xμL
3.25-x μL
total capacity
12.5μL ...
Embodiment 2
[0064] BRCA1 / 2 gene mutations were detected in 20 patients with breast tumors.
[0065] A. Genomic DNA extraction from whole blood of patients
[0066] Genomic DNA was extracted from the patient's peripheral blood samples using a blood genomic DNA extraction kit (Tiangen, Cat. No. DP318-02), and the operation was as described in the instructions of the extraction kit. use Fluorescence Quantitative Instrument and Corresponding Quantification reagent (Invitrogen) was used for quantification. The quantitative results are shown in Table 2.
[0067] Table 2. Sample quantification results
[0068]
[0069]
[0070] B. The multiplex PCR method amplifies the target region of the sample genome.
[0071] 1). Prepare a sterile, nuclease-free 200 μL PCR tube and place it on ice; prepare the PCR reaction system as shown in the table below, and be careful to avoid cross-contamination.
[0072] components
volume
2×PCR mix
6.25 μL
specific primer ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com