Mdr1a/1b double-gene knockout method and application
A gene knockout and double gene technology, applied in the field of biomedicine, can solve the problem of late emergence of mice in gene knockout rat models
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
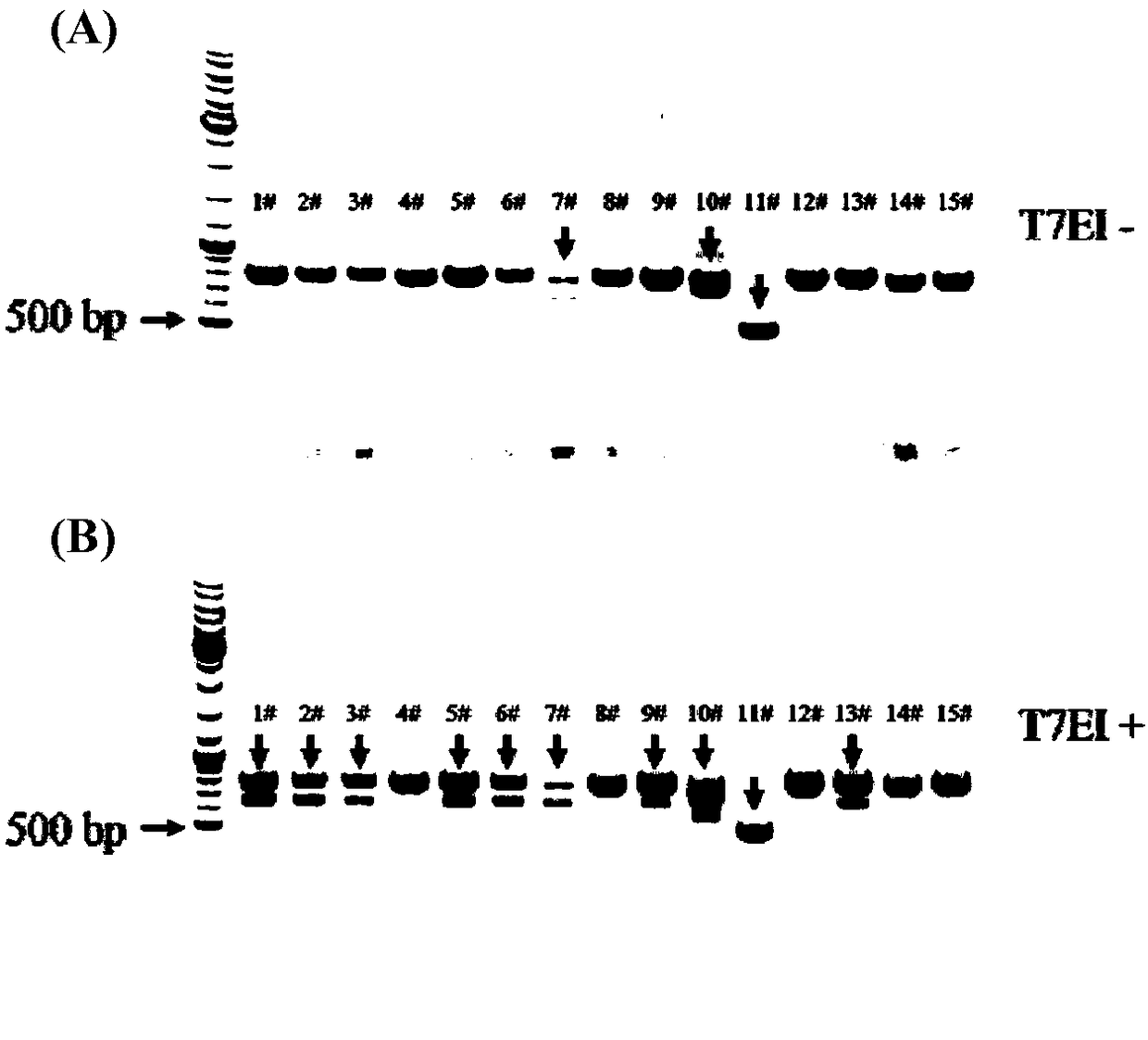
[0055] Example 1 Selection of Mdr1a and Mdr1b targets
[0056] The Mdr1a and Mdr1b gene sequences of the rat (Rattus norvegicus (Norway rat)) were respectively found in the NCBI database, and then the start codon and stop codon of the gene were found, and the exon region was marked. Since the first two exon sequences of Mdr1a and Mdr1b genes are short (the number of bases is less than 70bp), the target point is selected on the third exon. The third exon sequences of the two genes were successively input into the online target prediction website, and two targets with a length of 18bp were obtained.
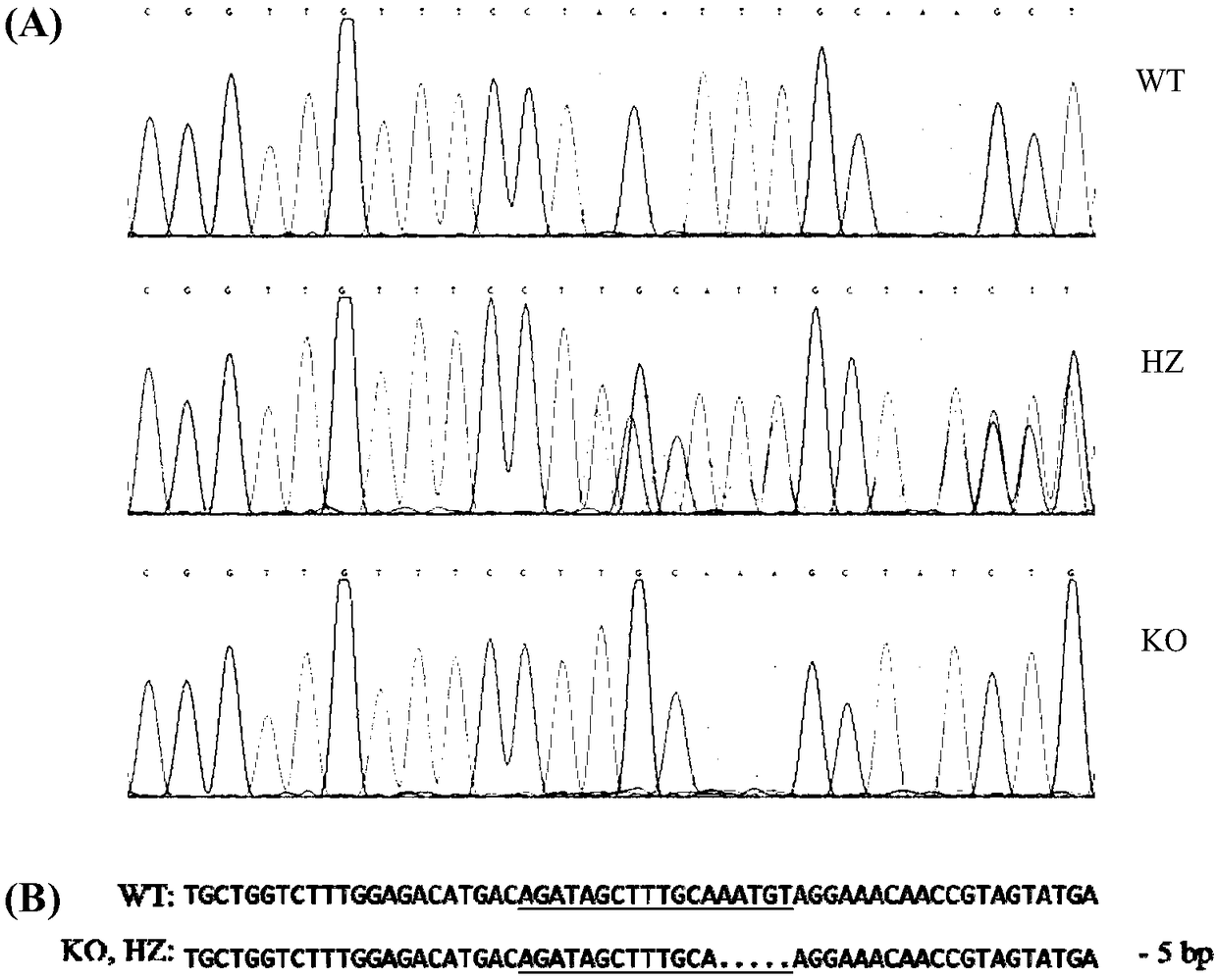
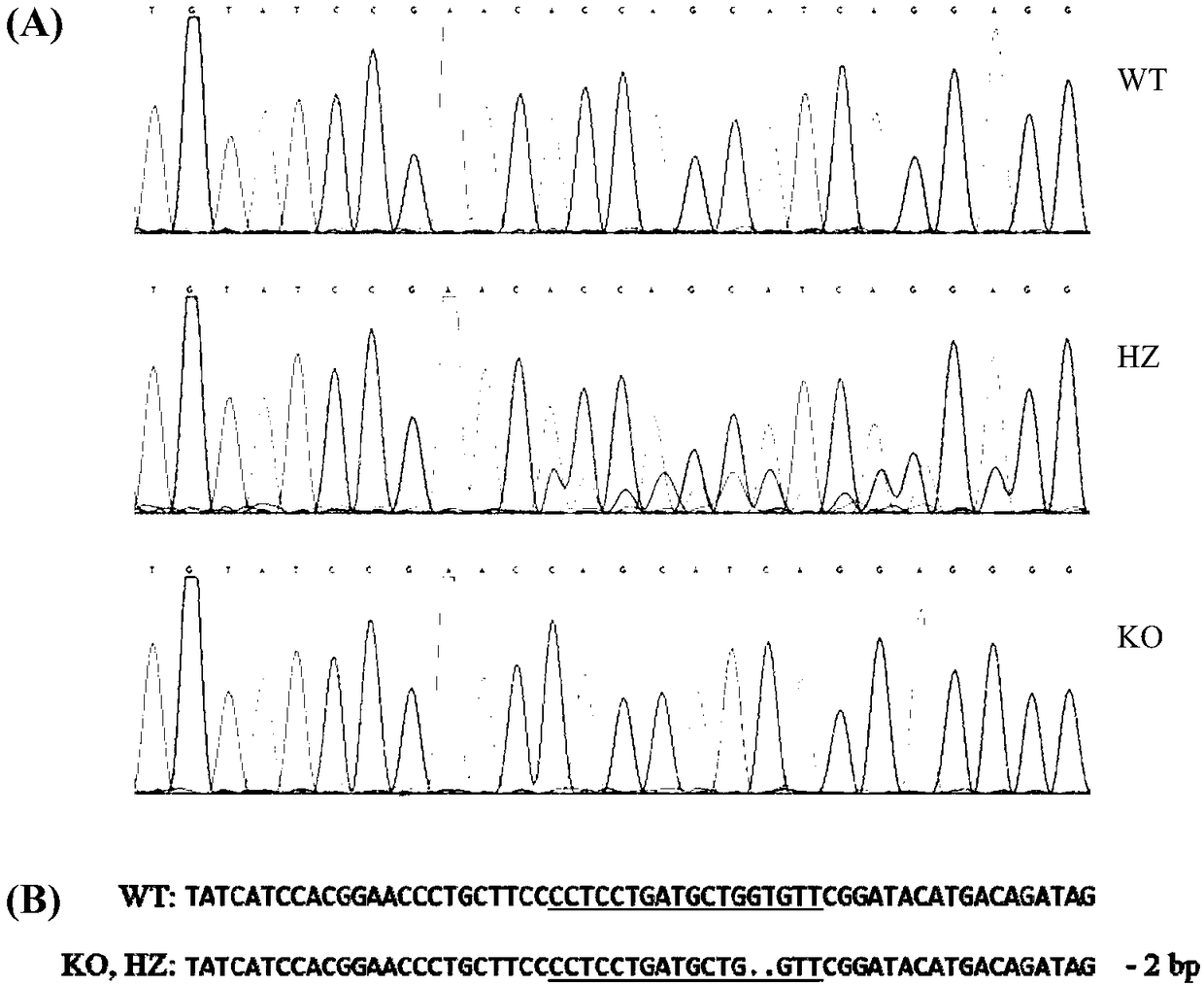
[0057] Among them, the online target prediction website is http: / / zifit.partners.org / ZiFiT / ChoiceMenu.aspx; the Mdr1a gene target sequence is 5'-AGATAGCTTTGCAAATGT-3' (SEQ ID NO.1); the Mdr1b gene target sequence is 5'-CCTCCTGATGCTGGTGTT-3' (SEQ ID NO. 2).
Embodiment 2
[0058] Synthesis and extraction of embodiment 2 sgRNA
[0059]First, synthesize a 60bp Oligo fragment, which includes the knockout target and T7 promoter sequence; then, using the Oligo fragment as a template, synthesize a complete sgRNA double-stranded template with a length of 130bp by overlapping PCR reaction, and pass The sgRNA double-stranded template was extracted and separated by the method of phenol-chloroform extraction; finally, the sgRNA double-stranded template was transcribed in vitro with the T7 in vitro transcription kit, and the transcription product was extracted and separated by the method of phenol-chloroform extraction to obtain sgRNA.
[0060] Among them, the Oligo fragment sequence containing the Mdr1a gene knockout target is 5'-GATCACTAATACGACTCACTATAGG AGATAGCTTTGCAAATGT GTTTTAGAGCTAGAAAT-3'(SEQ ID NO.3); the sequence of the Oligo fragment containing the Mdr1b gene knockout target is 5'-GATCACTAATACGACTCACTATAGG CCTCCTGATGCTGGTGTT GTTTTAGAGCTAGAAAT-3'...
Embodiment 3
[0061] Example 3 sgRNA and Cas9 mRNA co-injection and embryo transfer
[0062] (1) Preparation of pseudopregnant mice. Select robust male SD rats over 8 weeks old for sterilization, and then select robust female SD rats 7-8 weeks old to mate with the above-mentioned sterile male mice. After mating, the female mouse will show signs of pregnancy, which is the desired pseudopregnant mouse.
[0063] (2) Collection and microinjection of fertilized eggs. Strong, 6-7-week-old female SD rats were selected for superovulation, and then mated with healthy, normal-born male rats. On the second day of mating, the female mice were sacrificed, and the fertilized eggs were collected for later use. Before microinjection, the collected fertilized eggs were incubated in embryo medium at 37°C CO 2 After culturing in an incubator for 3-4 hours, the sgRNAs of Mdr1a and Mdr1b were mixed with Cas9 mRNA and then injected into the cytoplasm of fertilized eggs. Among them, the concentration of the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com