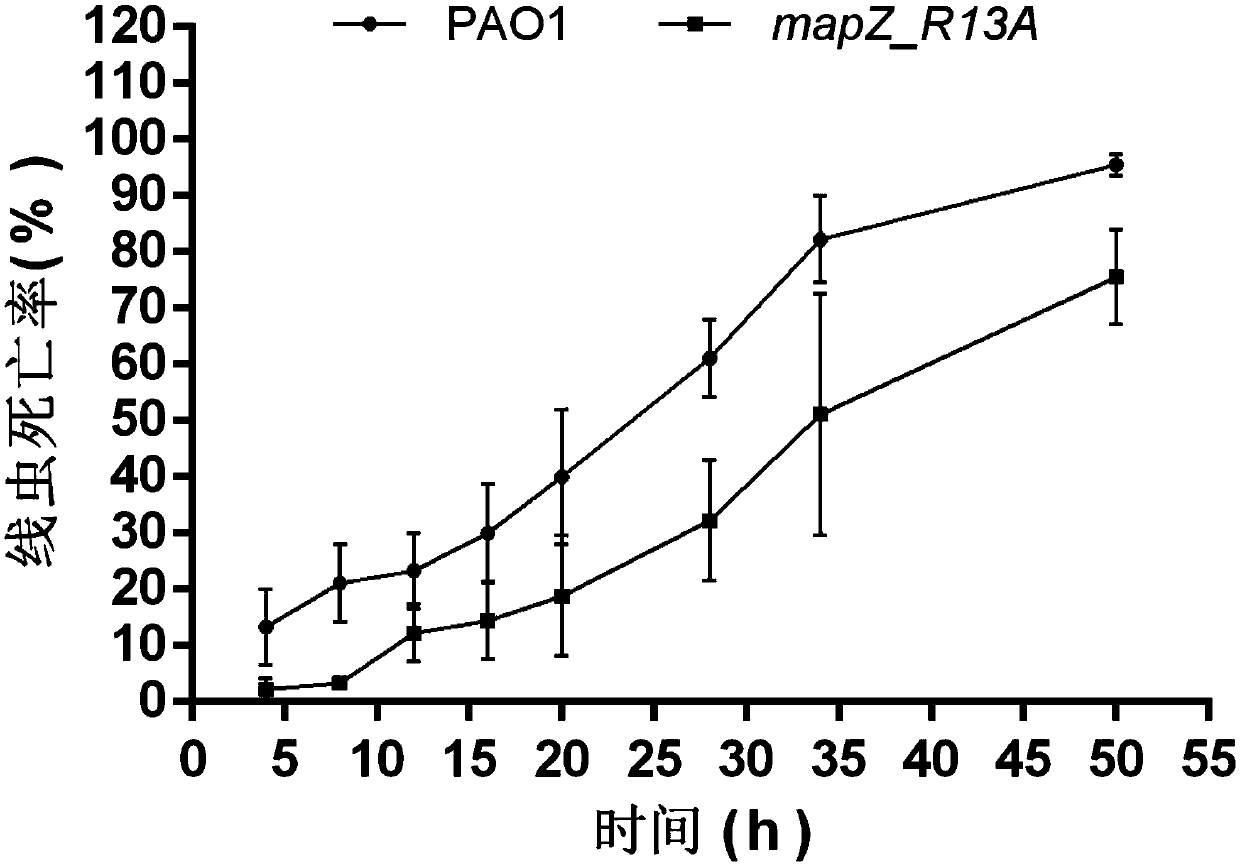
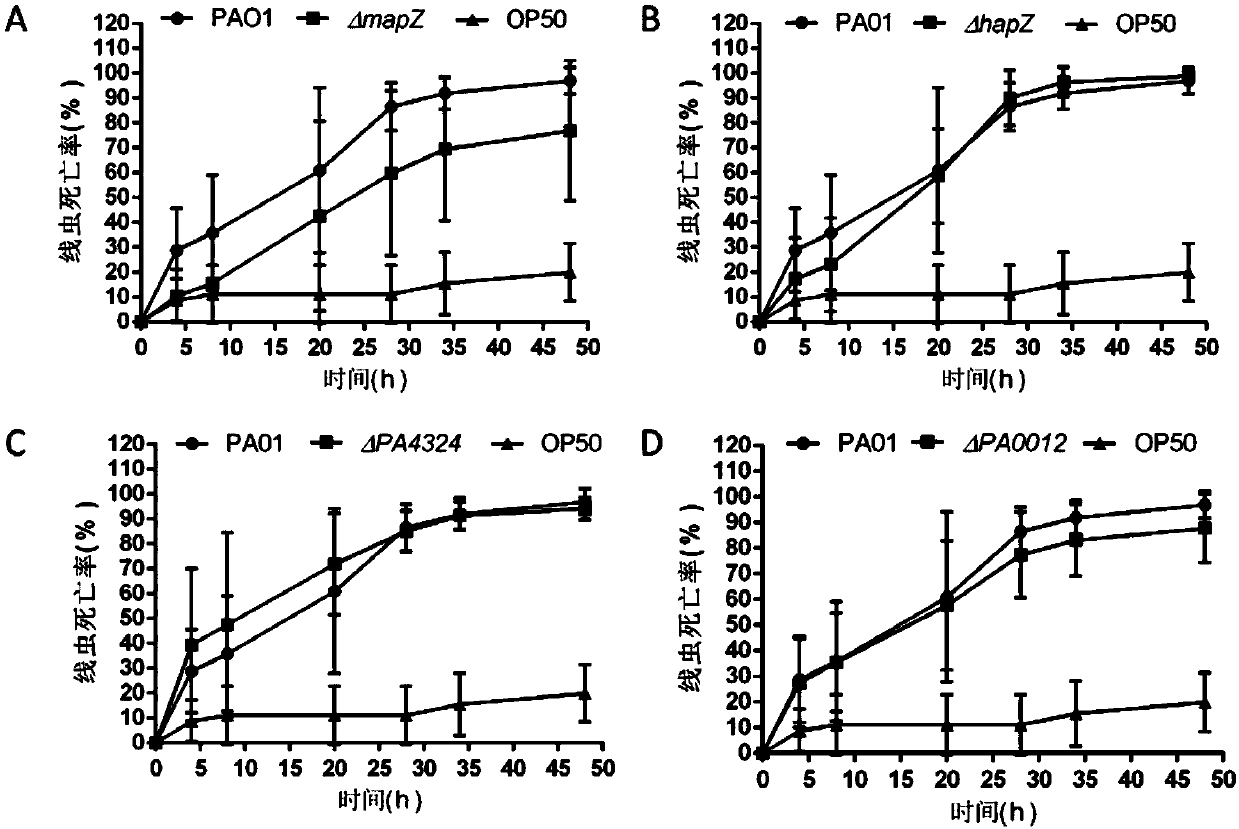
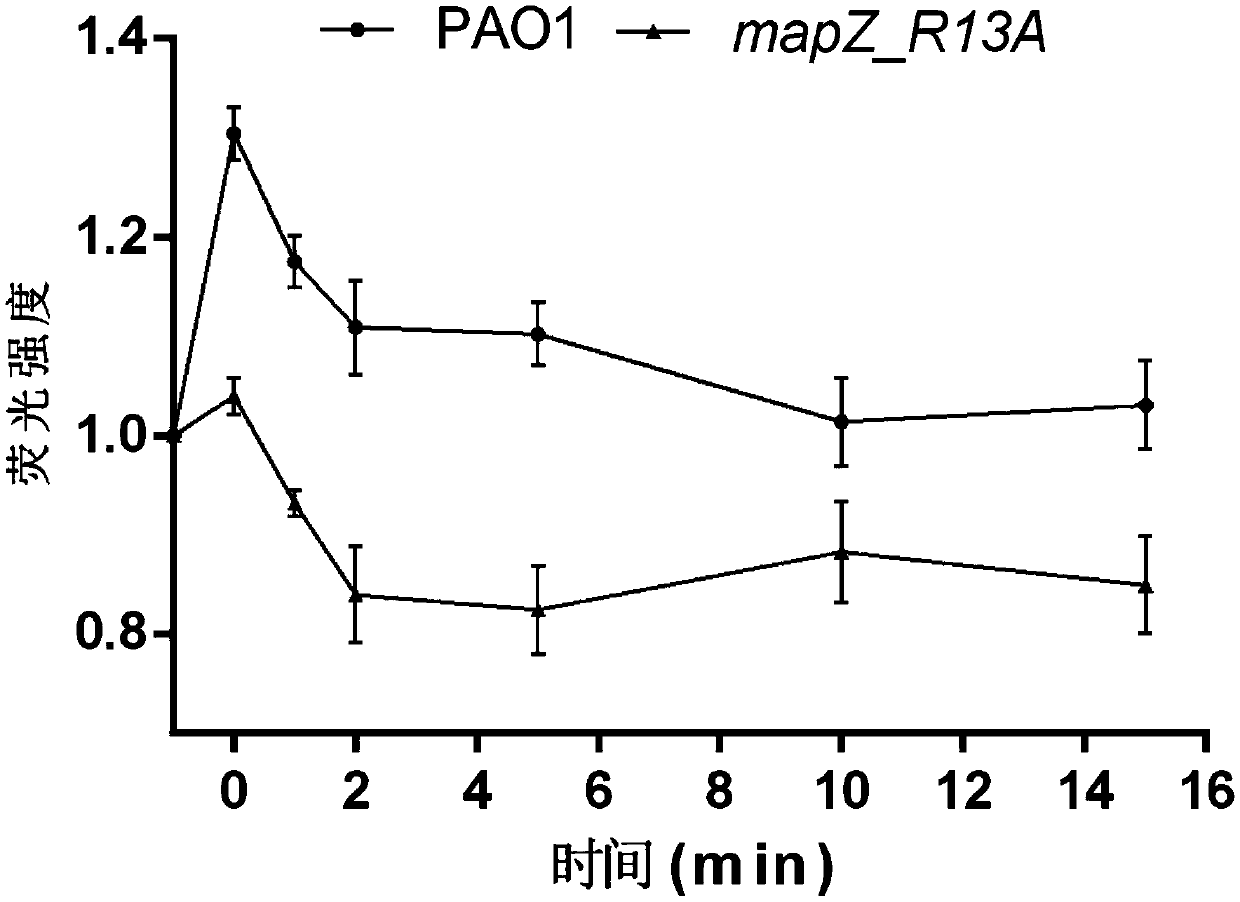
Application of PA4608 protein as target in preparing antibacterial drugs
A technology of antibacterial drugs and targets, applied in the field of application in the preparation of antibacterial drugs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1 Construction of mapZ_R13A mutant strain
[0036] In this example, the mapZ_R13A mutant strain was constructed by homologous exchange recombination technology.
[0037] 1. A point mutation is carried out at the 13th arginine (R13) of the c-di-GMP binding site of the protein encoded by the gene PA4608, and the MapZ protein (its amino acid sequence is shown in SEQ ID NO.1, and its nucleotide sequence is shown in SEQ ID NO.2) and MapZ R13A The amino acid sequence of the protein is compared as figure 1 As shown, the gene synthesis method was used to synthesize mapZ containing upper and lower homology arms R13A A gene fragment, the nucleotide sequence of which is shown in SEQ ID NO.3.
[0038] 2. Design primer F (CGCGGATCCTGGCGAAGATCGAGCGGCCG, SEQ ID NO.6) and primer R (CCCAAGCTTTGCCGAGCGGCCAGGAGCGC, SEQ ID NO.7), and use the DNA obtained in step 1 as a template for PCR amplification. The reaction system is shown in Table 1:
[0039] Table 1 PCR reaction system
...
Embodiment 2
[0045] Example 2 Construction of ΔmapZ mutant strain
[0046] In this example, the ΔmapZ mutant strain was constructed by homologous exchange recombination technology.
[0047] 1. To knock out the mapZ gene, design primers P1 (CGCGGATCCAAGTGCGCCCGGTCCTTGGCTT, SEQ ID NO.8) and P2 (CCCTCTTCGAGTGACCCGCCTCTCATGTCGCGATCCCTTGGTGC, SEQ ID NO.9), and use PAO1 genomic DNA as a template to amplify the homology arm on the mapZ gene. The reaction system is as follows Shown in Table 2; Design primers P3 (GCACCAAGGGATCGCGACATGAGAGGCGGGTCACTCGAAGAGGG, SEQ ID NO.10) and P4 (CCCAAGCTTCCGTACTGTATTTCGAGGGCGA, SEQ ID NO.11), using PAO1 genomic DNA as a template to amplify the lower homology arm of the mapZ gene, the reaction system is shown in Table 3 Show.
[0048] Table 2 PCR reaction system
[0049]
[0050] Table 3 PCR reaction system
[0051]
[0052] The PCR amplification conditions were: pre-denaturation at 98°C for 30s, denaturation at 98°C for 10s, annealing at 65°C for 30s, ext...
Embodiment 3
[0059] Example 3 Construction of ΔhapZ mutant strain
[0060] In this example, the ΔhapZ mutant strain was constructed by homologous exchange recombination technology.
[0061] 1. Knockout the hapZ gene (ie PA2799), design primers P5 (CGCGGATCCAAAGTACGATGATCGGCGTTTC, SEQ ID NO.13) and P6 (CGCCTTTCGTCGTTTCAGTGGAGCTCGATGGGCTGCATGGGCTG, SEQ ID NO.14), and use PAO1 genomic DNA as a template to amplify the homology of the hapZ gene Arm, the reaction system is the same as in Table 2; primers P7 (CAGCCCATGCAGCCCATCGAGTCCCACTGAAACGACGAAAGGCG, SEQ ID NO.15) and P8 (CCCAAGCTTTTGCAGGCCAAGTTCCCAATG, SEQ ID NO.16) were designed to amplify the lower homology arm of the hapZ gene with PAO1 genomic DNA as a template, and the reaction system is the same as in the table 3.
[0062] The PCR amplification conditions were: pre-denaturation at 98°C for 30s, denaturation at 98°C for 10s, annealing at 65°C for 30s, extension at 72°C for 30s, a total of 30 cycles, extension at 72°C for 5min, and stor...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com