Gene editing system for preparing allotransplantable T cells
A technology of allotransplantation and gene editing, applied in plant gene improvement, genetic engineering, introduction of foreign genetic material using vectors, etc., can solve the problems of long cycle, high cost and complex construction process.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1 gRNA preliminary screening
[0047] 1. Preparation of gRNA targeting TCRA gene
[0048] (1) The gRNA sequence designed according to the sequence of the TCRA gene (including the constant region of the α chain and the β chain):
[0049] The target sequence corresponding to the gRNA of the CRISPR / SpCas9 system is shown in the table below:
[0050] Table 1 Target sequence corresponding to the gRNA of the CRISPR / SpCas9 system for the TCRA gene.
[0051]
[0052]
[0053] The target sequence corresponding to the gRNA of the CRISPR / SaCas9 system is shown in the table below:
[0054] Table 2 The target sequence corresponding to the gRNA of the CRISPR / SaCas9 system for the TCRA gene.
[0055]
[0056]
[0057] (2) Synthesize the sense strand and antisense strand of the target sequence corresponding to the gRNA respectively (add cacc at the 5'-end of the sense strand, if the first nucleotide at the 5'-end of the sense strand is not guanine G, then add cac...
Embodiment 2
[0098] Example 2 Screening for gRNA combinations of TCRA genes
[0099] In Example 1, through T7E1 enzyme digestion and agarose electrophoresis analysis, the gRNA with higher mutation efficiency screened out was combined to perform mutation operation on the TCRA gene. The specific steps are as follows:
[0100] 1. The gRNA combination is shown in the table below:
[0101] Table 4 The target sequence corresponding to the gRNA combination of the CRISPR / SaCas9 system for the TCRA gene:
[0102]
[0103] The plasmids prepared in 3(6) of Example 1 were co-transfected into HEK293T cells according to the above combination method, and the transfection method was referred to 4(1)-4(3) of Example 1;
[0104] 48 hours after transfection, amplify and purify the target fragment containing the target site according to the method of 5(1)~5(4) in Example 1;
[0105] Using DL2000 (TAKARA) as a reference, the length of the above PCR product was detected by 2% agarose gel electrophoresis. ...
Embodiment 3
[0111] Example 3 CRISPR / Cas9-mediated site-directed homologous recombination of TCRA gene
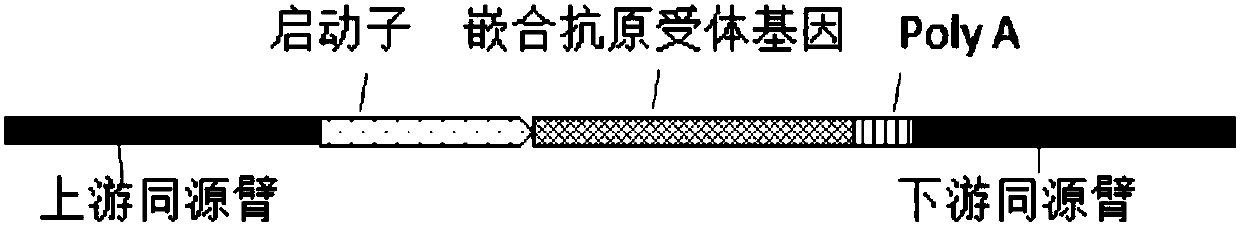
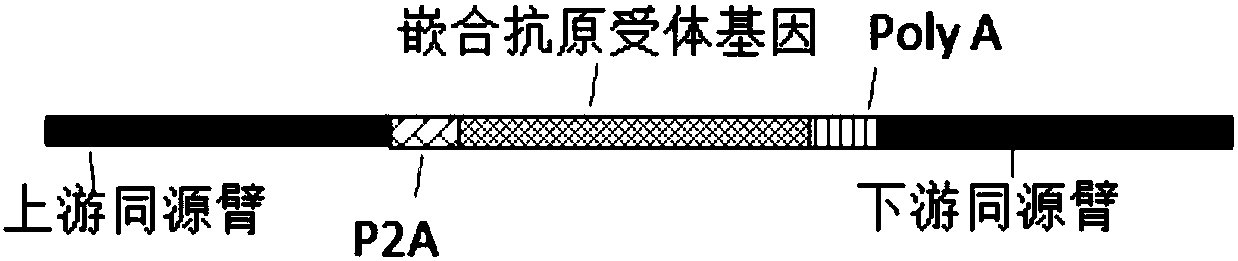
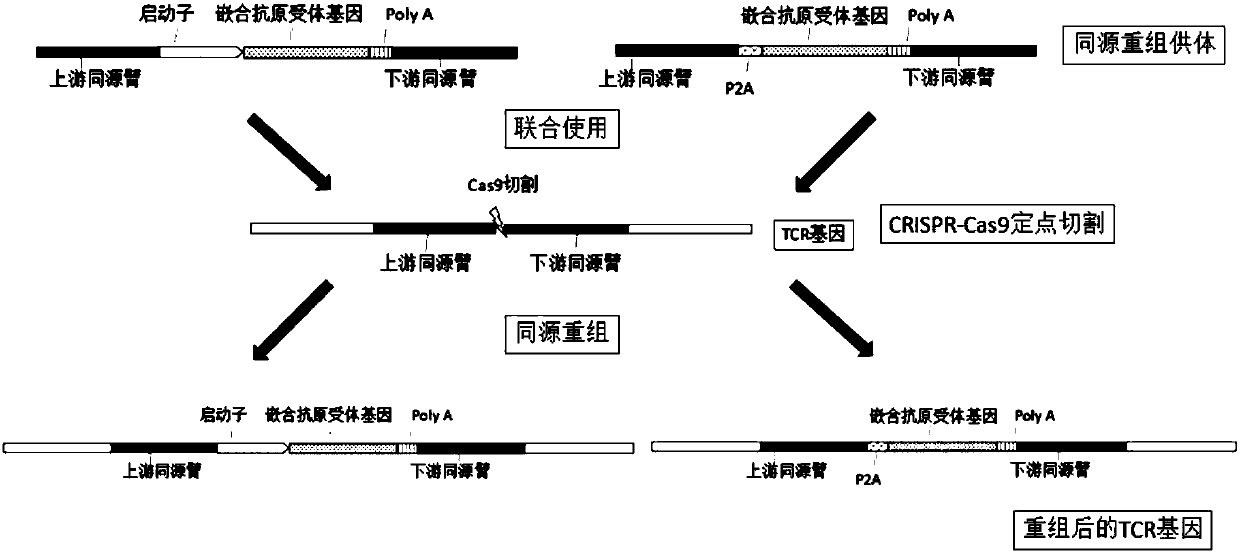
[0112] The gRNA or gRNA combination with higher mutation efficiency screened in Example 1 or Example 2 is used for CRISPR / Cas9-mediated integration and insertion of a foreign DNA sequence at a specific site in the TCRA gene (Cas9 cleavage site) . The following examples take gRNA target sequence SEQ ID NO: 3 as an example, and similarly can demonstrate the implementation effect of other gRNA or gRNA combinations.
[0113] 1. Homologous recombination donor vector construction
[0114] (1) Use the HEK293T cell genome as a template, use SEQ ID NO: 65 and SEQ ID NO: 66 as primers to amplify the upstream homology arm, and use SEQ ID NO: 67 and SEQ ID NO: 68 as primers to amplify the downstream homology arm , The PCR product was detected by 1% agarose gel electrophoresis, and the target fragment was recovered by cutting the gel;
[0115] Table 6 Amplification primers 65, 66, 67, 68 of the u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com