Method for detecting GII type norovirus in marine food
A virus and food technology, applied in biochemical equipment and methods, microbiological determination/inspection, DNA/RNA fragments, etc., can solve the problem of norovirus highly infectious, false negative, mutant strain or genotype detection methods. and other problems, to achieve the effect of wide application prospects, low false positive and false negative, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1, real-time fluorescent PCR primer design
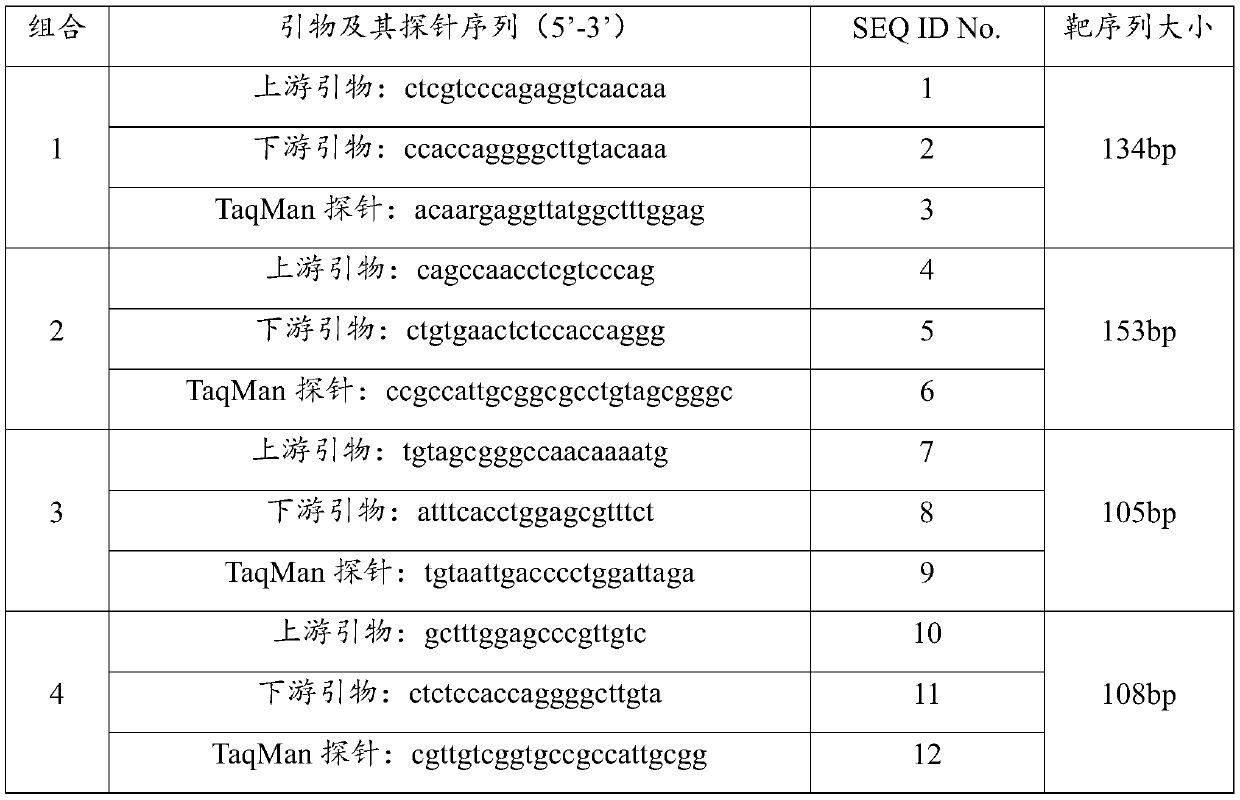
[0032] The sequence information of all GⅡ norovirus VP1 genes was retrieved from NCBI. After sequence analysis and comparison, the highly conserved region of GⅡ norovirus was found, 8 pairs of primers and probes were designed, and BLAST sequence comparison was performed. , screen out 4 pairs of primers and their probe combinations with better specificity, and then carry out the biosynthesis of the sequence, in which, the 5' end of each probe is labeled with a fluorescent reporter group FAM, and the 3' end is labeled with a quencher group BHQ1, each combined sequence is shown in Table 1.
[0033] Table 1. Sequences of primers and their probe combinations
[0034]
Embodiment 2
[0035] Example 2. Specificity of real-time fluorescent PCR detection of GⅡ type norovirus
[0036] Use 4 pairs of primers and probe combinations thereof shown in Table 1 in Example 1 to carry out real-time fluorescent PCR detection of oyster samples infected by the following different viruses: rotavirus, astrovirus, hepatitis A virus, different subtypes GI. GI Noroviruses from 1 to 9, GII Noroviruses of different subtypes GII.1 to 21, GIII to VII Noroviruses, each sample was repeated three times, and the results were as follows:
[0037] The detection results of blank control and negative control using different primers and their probe combinations were all negative;
[0038] The primers and probe combinations 1 and 2 in Table 1 are positive for all different subtypes of GⅡ norovirus strain-infected samples, while other samples are negative;
[0039] The primers and their probe combinations 3 in Table 1 were positive for all different subtypes of GⅡ norovirus-infected samples...
Embodiment 3
[0053] Example 3, Sensitivity of real-time fluorescent PCR detection of GⅡ type norovirus
[0054] Measure the value of OD260 / OD280 of the DNA solution of the sample identified as GⅡ type norovirus positive in Example 2, and calculate the DNA concentration, use RNase-free Water to dilute to a certain concentration, and then serially dilute to a concentration of 1 pg respectively / μL, 0.1pg / μL, 0.01pg / μL, 0.005pg / μL, 0.001pg / μL of different standard samples.
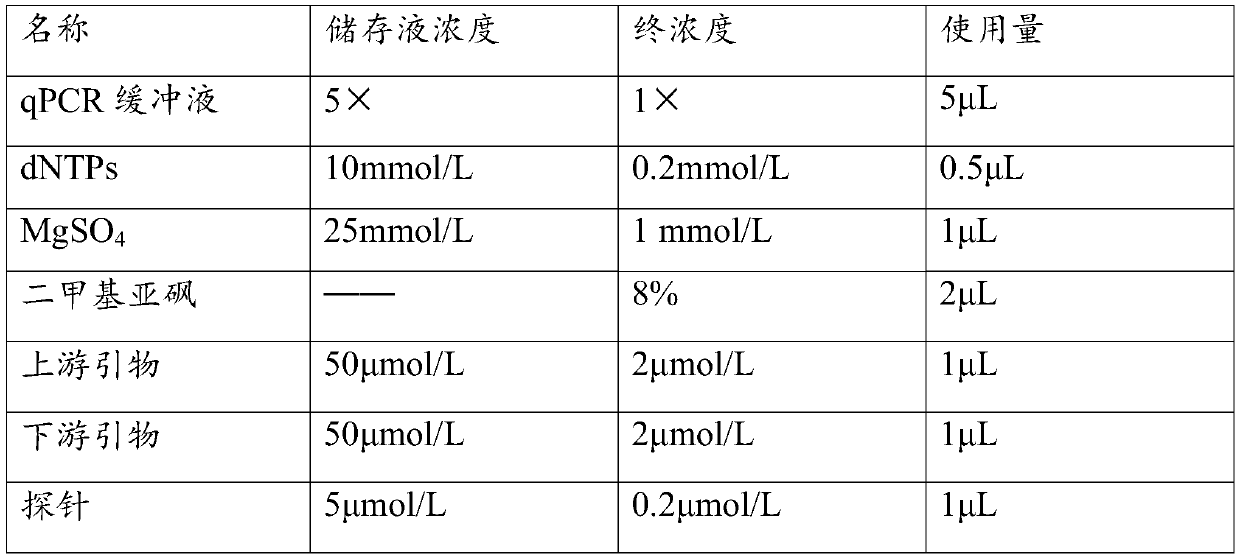
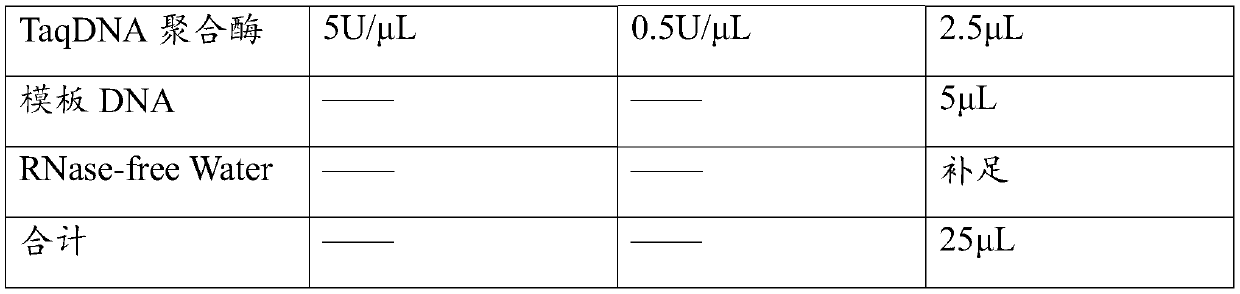
[0055] Using the above-mentioned different standard samples as templates, the primers and their probe combinations 1 and 2 in Table 1 were used respectively, and the detection was carried out according to the method of step 2 in the real-time fluorescent PCR detection described in Example 2.
[0056] Results: The primer and probe combination 1 in Table 1 is positive when the concentration of the standard sample is 0.005pg / μL and above, and negative when the concentration of the standard sample is 0.001pg / μL;
[0057] The...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com