Method for preparing 10-hydroxyl-2-decenoic acid with decylic acid as raw material and by utilizing escherichia coli engineering bacteria
A technology of Escherichia coli and engineering bacteria, which is applied in the field of preparing 10-hydroxy-2-decenoic acid, can solve the problems of related technologies that have not been reported, and achieve the effect of improving conversion rate and efficient assembly
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
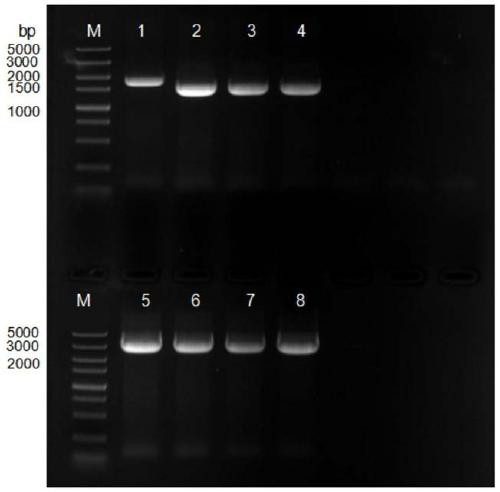
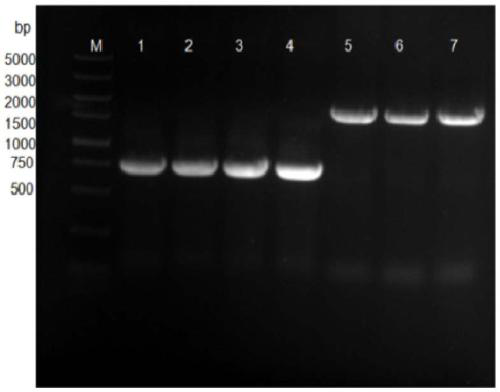
[0072] Example 1: PCR amplification of Escherichia coli acyl-CoA thioesterase gene ydiI, MCAD, FadK, P450 fusion enzyme.
[0073] According to Escherichia coli ester acyl-CoA thioesterase gene P450 fusion enzyme, FadK, MCAD, ydiI design PCR amplification primer, the nucleotide sequence of upstream primer is shown in SEQ ID NO.1,3,5,7 respectively, downstream The nucleotide sequences of the primers are shown in SEQID NO.2, 4, 6, and 8;
[0074] Wherein, the nucleotide sequences of the Escherichia coli acyl-CoA thioesterase gene P450 fusion enzyme, FadK, MCAD, and ydiI are shown in SEQ ID NO.9, 10, 11, and 12 respectively, and the expressed small molecule thioesterase The amino acid sequence is shown in SEQ ID NO.13, 14, 15 and 16.
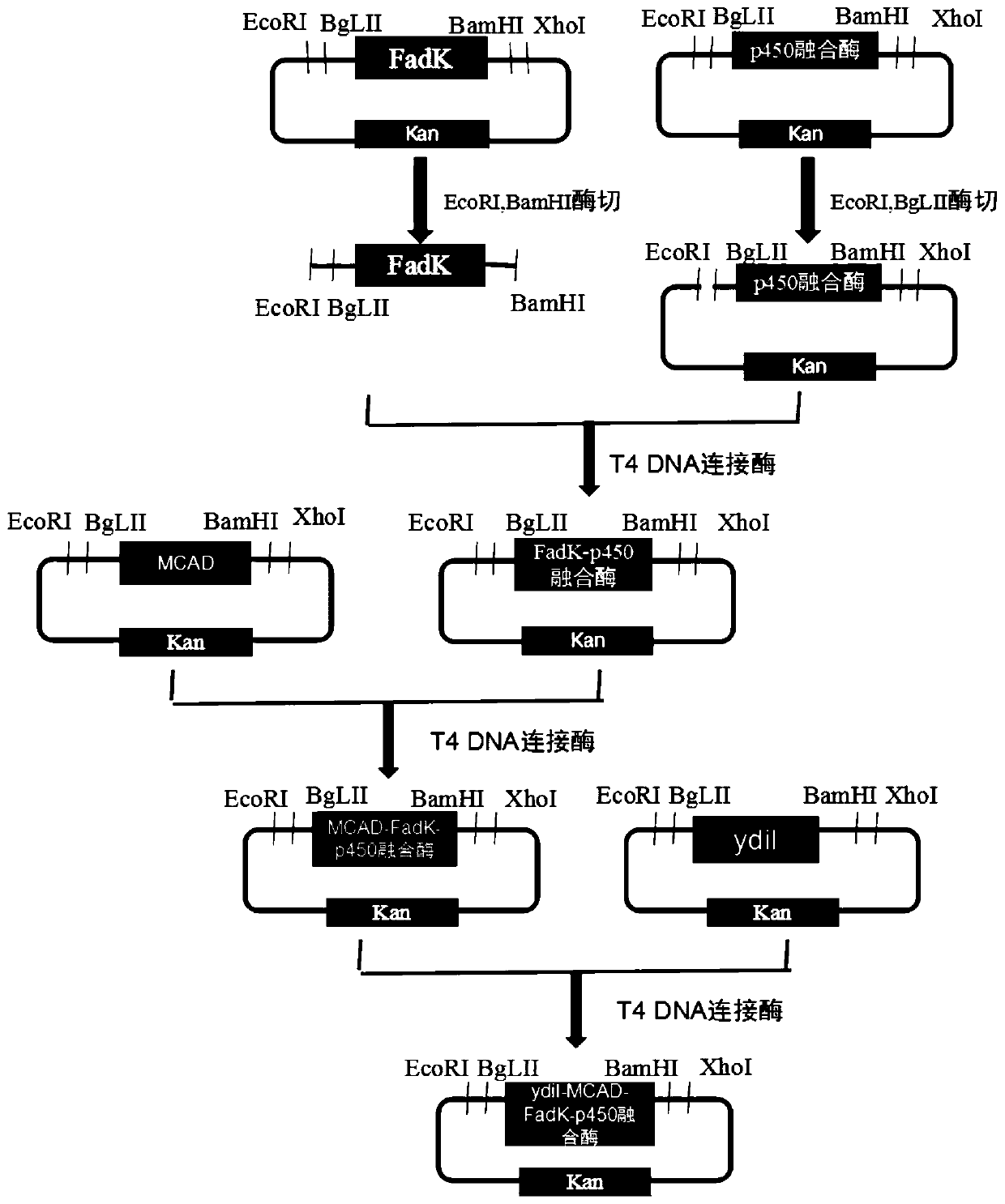
[0075] The construction contains recombinant plasmid pBbB5K-P450 fusion enzyme, comprising the following steps:
[0076] The codon-optimized fusion enzyme gene of alkane hydroxylase CYP153A of Marinobacter aquaeolei and P450 NADH reductase of Baci...
Embodiment 2
[0101] Embodiment 2: Construction of recombinant plasmid pBbB5K-ydiI, pBbB5K-MCAD, pBbB5K-FadK, pBbB5K-P450 fusion enzyme
[0102] The PCR product recovered in Example 1 and the double enzyme digestion reaction of the pBbB5K-GFP plasmid vector, the reaction system is as follows:
[0103]
[0104] Reaction conditions: react at 37°C for 1.5h.
[0105] PCR products and plasmid vectors were digested and purified by 1% agarose gel electrophoresis, and the target fragments were recovered using a DNA gel recovery kit.
[0106] Ligate the digested PCR product with the plasmid vector that has also been digested, and the ligation reaction system is as follows:
[0107]
[0108] Mix the above ligation reaction system thoroughly and centrifuge for a few seconds, collect the drop on the tube wall to the bottom of the tube, and ligate at 22°C for 10 minutes to obtain recombinant plasmids pBbB5K-ydiI, pBbB5K-MCAD, pBbB5K-FadK, pBbB5K-P450 fusion enzyme.
Embodiment 3
[0109] Embodiment 3: Transformation of recombinant plasmid pBbB5K-ydiI, recombinant plasmid pBbB5K-MCAD, recombinant plasmid pBbB5K-FadK, recombinant plasmid pBbB5K-P450 fusion enzyme:
[0110] (1) Preparation of Competent Cells
[0111] ①Pick a single colony of Escherichia coli BL21 (DE3) (or pick a preserved strain) and inoculate it into 10ml of liquid LB medium, culture overnight at 37°C and 210rpm;
[0112] ② Inoculate 5ml of the bacterial solution into 500ml of LB medium, and cultivate at 37°C and 210rpm until the OD600 of the bacterial solution is about 0.375;
[0113] ③ Place the bacterial solution on the ice-water mixture for 10 minutes, and pre-cool the 50ml centrifuge tube at the same time;
[0114] ④ Transfer the bacterial solution to a centrifuge tube, centrifuge at 3700 rpm for 10 minutes at 4°C to collect the bacterial cells;
[0115] ⑤Add 10mL of pre-cooled 0.1M CaCl2 solution to each centrifuge tube, resuspend the bacteria, then add 30mL of pre-cooled 0.1M Ca...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com