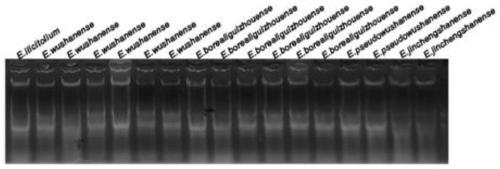
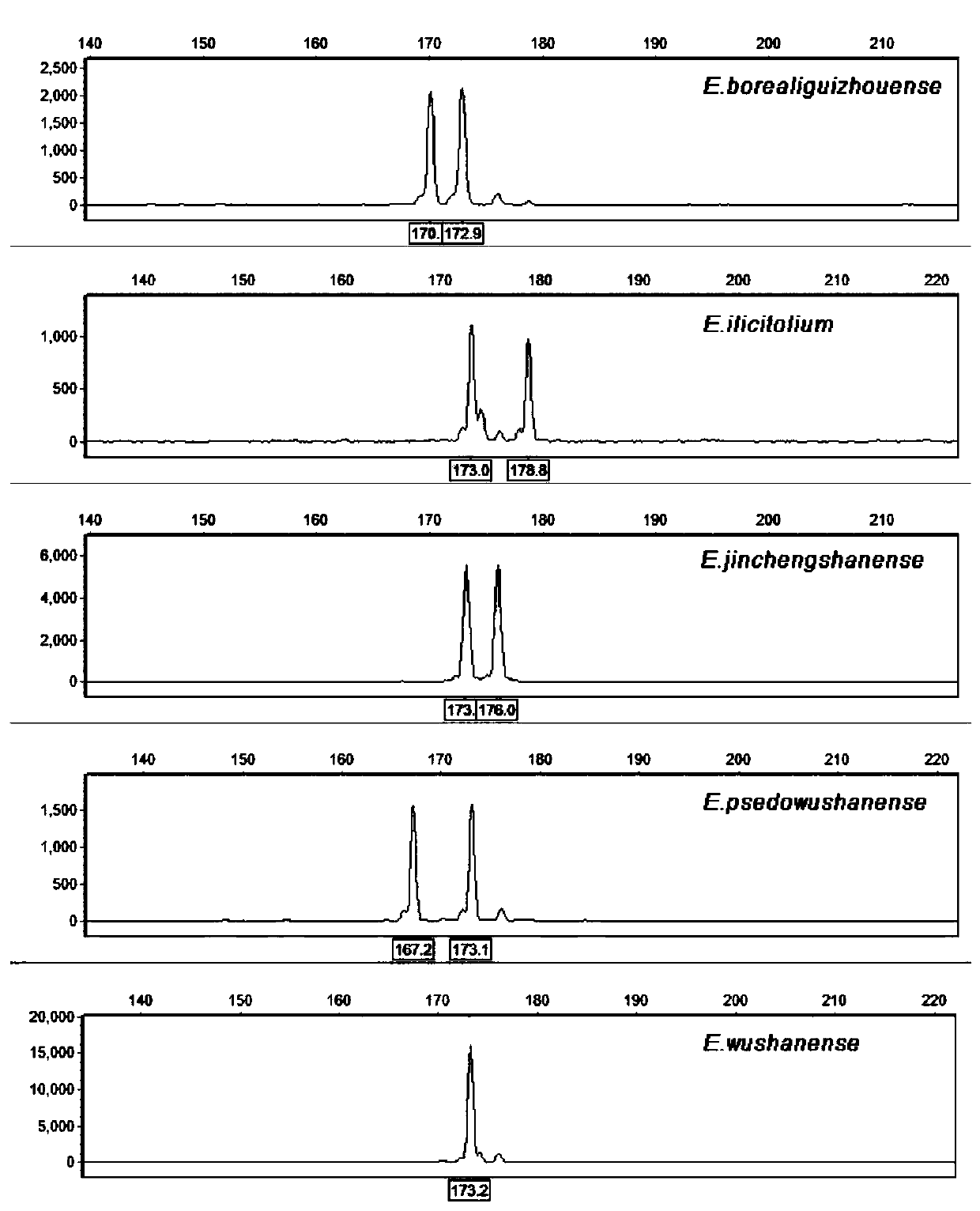
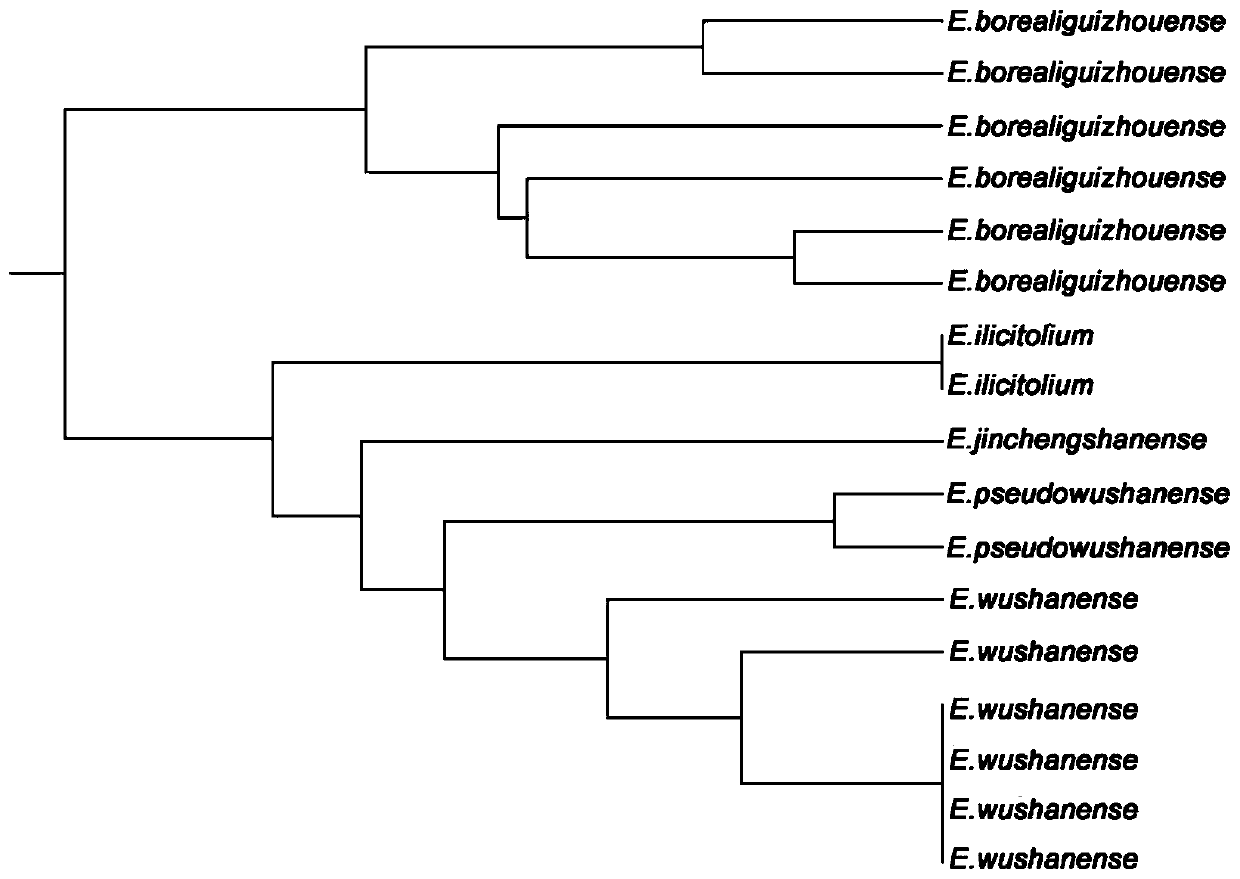
EST-SSR molecular identification method for E.wushanense and easily mixed species thereof
A technology of molecular identification and Epimedium, which is applied in biochemical equipment and methods, microbiological determination/inspection, DNA/RNA fragments, etc., can solve problems such as differences in the composition and content of medicinal ingredients, and achieve rapid and accurate identification, expansion Stable results and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Embodiment 1: Epimedium SSR primer design
[0028] Use the MicroSatellite (MISA) software to perform SSR searches on the Unigenes in the Epimedium transcriptome above 1kb in the database with the repeat times of 10, 6, and 5 for single nucleotides, dinucleotides, and trinucleotides; The Primer 6 software designed the target primers for the searched SSR sites. The design criteria were: the primer length was 18-26bp, the annealing temperature was 50-65°C, and the length of the final product of PCR amplification was 100-350bp. 5471 pairs of primers were synthesized from the SSR sites compared with the redundant protein database, and each pair of primers consisted of an upstream primer F and a downstream primer R.
Embodiment 2
[0029] Example 2: Extraction and detection of sample genomic DNA
[0030] The extraction and detection of genomic DNA from the leaves of Epimedium wushanense and its four easily mixed species, the specific steps are as follows:
[0031] A. Select the young leaves of the sample to be tested, remove the surface dust with 75% alcohol, preheat CTAB buffer at 65°C, and pre-cool isopropanol at -20°C;
[0032] B. Take 0.1g of dried plant leaves, freeze them in liquid nitrogen, grind them into powder, put them into 2mL centrifuge tubes, add 1mL of CTAB buffer solution preheated to 65°C, mix them upside down, and incubate at 65°C for 30 minutes. Shake well every 10 minutes;
[0033] C. After incubation, centrifuge at 12000r / min for 10 min, take 900μL of supernatant, add an equal volume of chloroform-isoamyl alcohol (24:1 (V / V)), mix well and extract for 5min, centrifuge at 12000r / min 10min, take the supernatant;
[0034] D. Repeat step C, add an equal volume of isopropanol pre-coole...
Embodiment 3
[0040] Embodiment 3: PCR amplification
[0041] Using the synthesized 10 pairs of EST-SSR core primers and universal M13 primers, perform PCR amplification on the genomic DNA of Epimedium wushanense and its four easily mixed species. The reaction system is: 2×Taq PCR Master Mix reaction buffer 5 μL , 0.0125 μL and 0.25 μL of 10 μM upstream and downstream primers, 0.15 μL of M13-ROX / FAM / HEX / TAMRA adapter primer, 3 μL of diluted DNA template, and make up to 10 μL with sterile water; Denaturation for 4min, denaturation at 94°C for 30s, annealing temperature from 65°C to 50°C for a total of 15 cycles, each cycle lowering 1°C, then 22 cycles at annealing temperature of 50°C, extension at 50°C for 30s, final extension at 72°C for 42s , stored at 16°C.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com