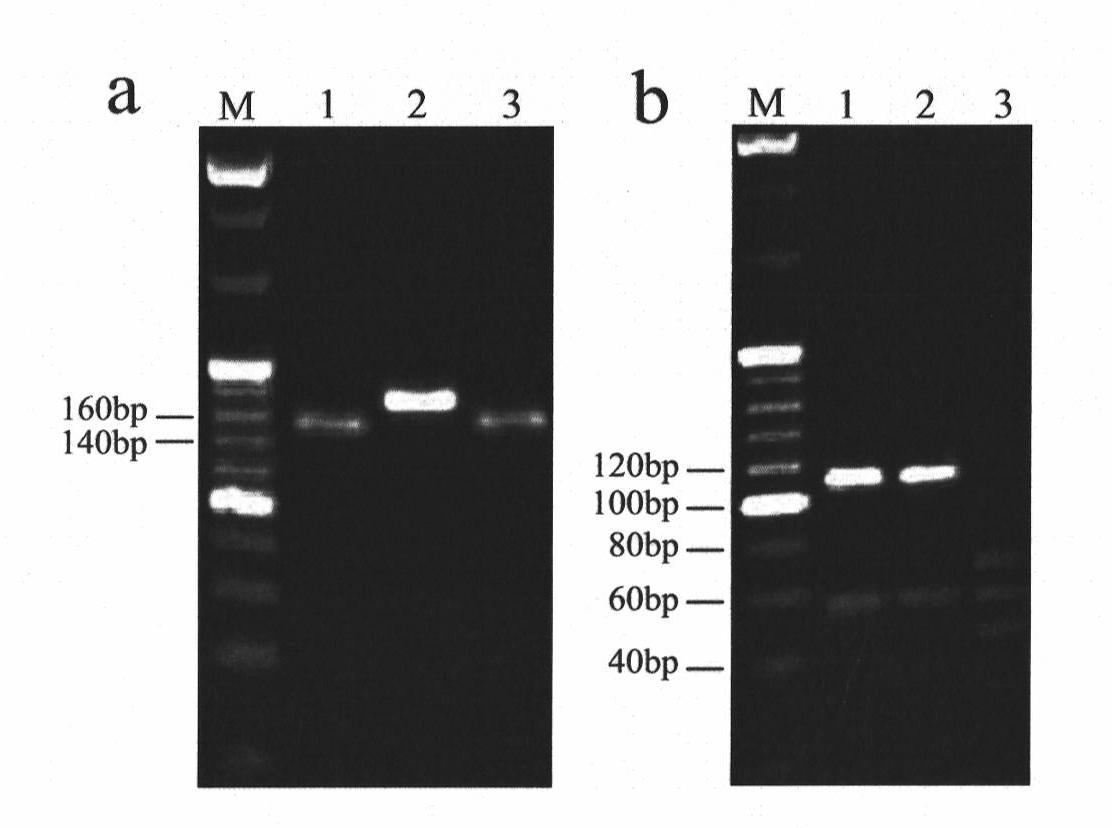
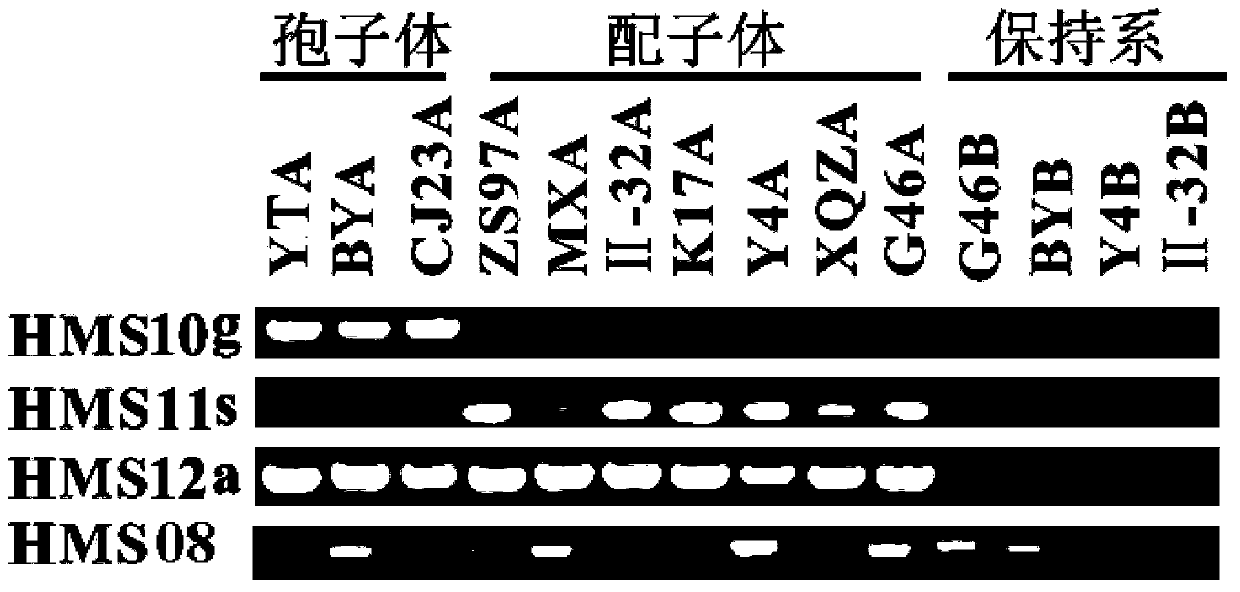
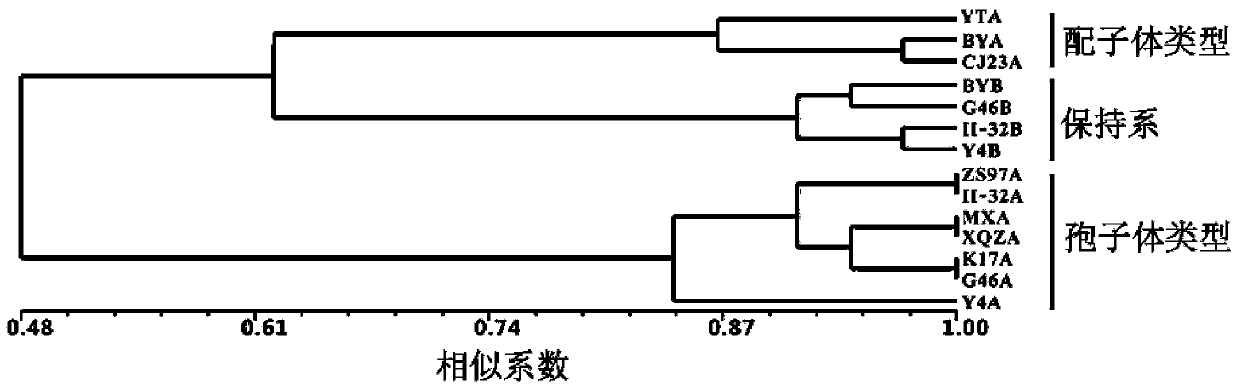
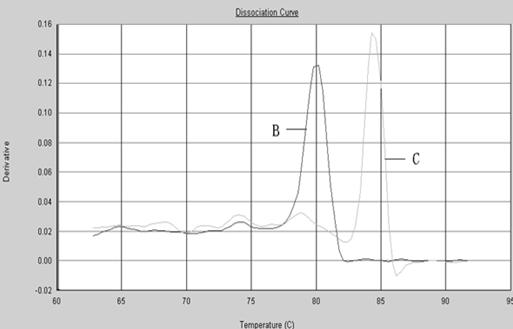
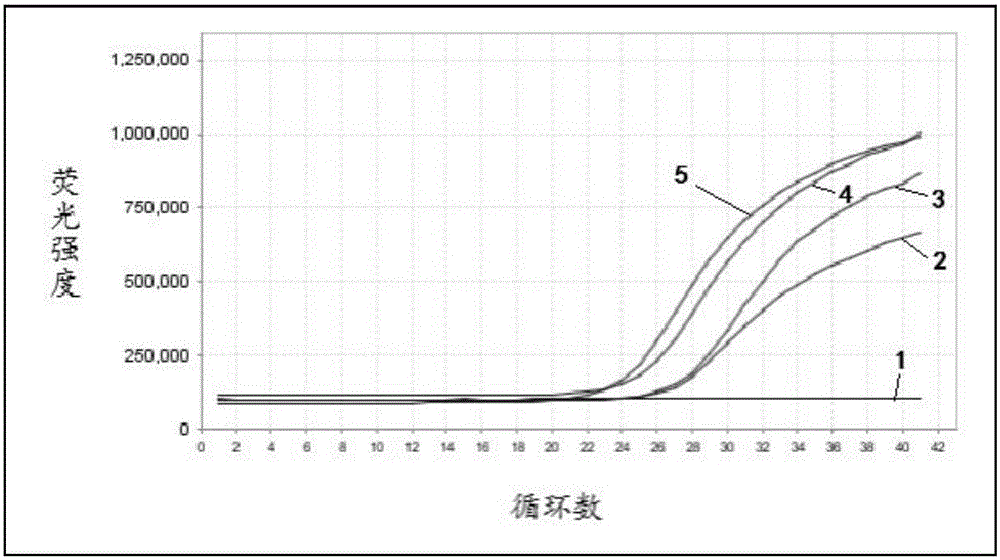
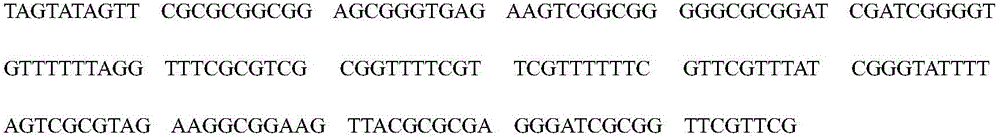
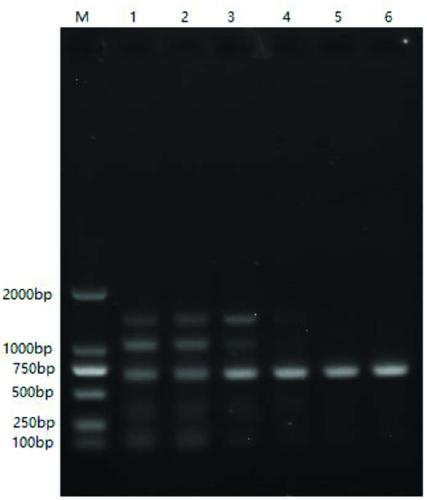
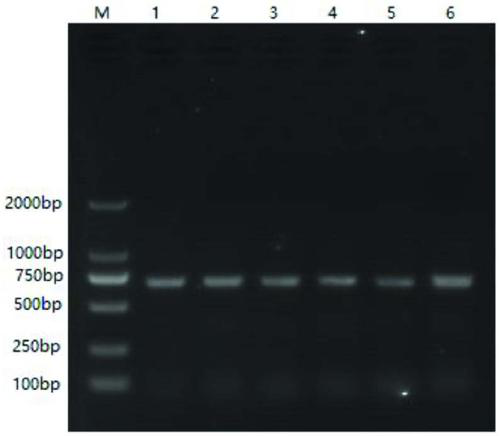
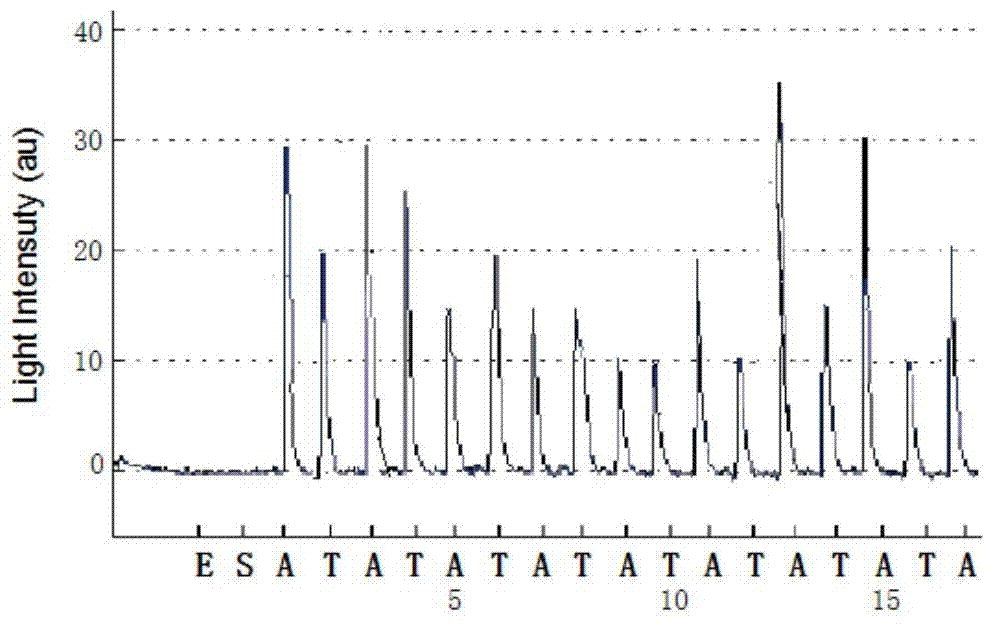
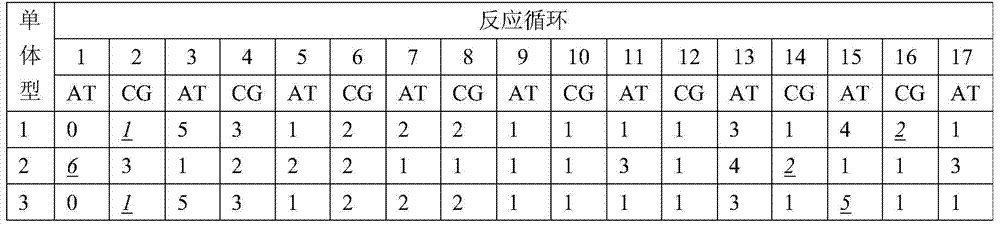
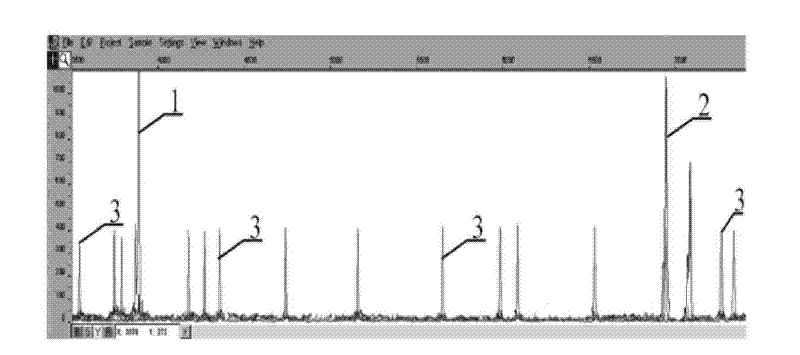
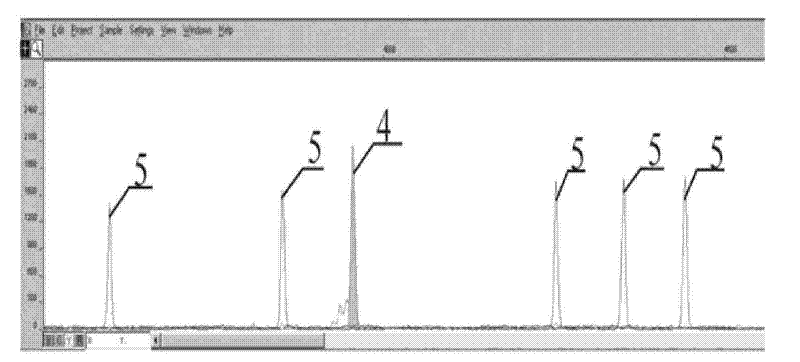
Patents
Literature
32 results about "Analysis pcr" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
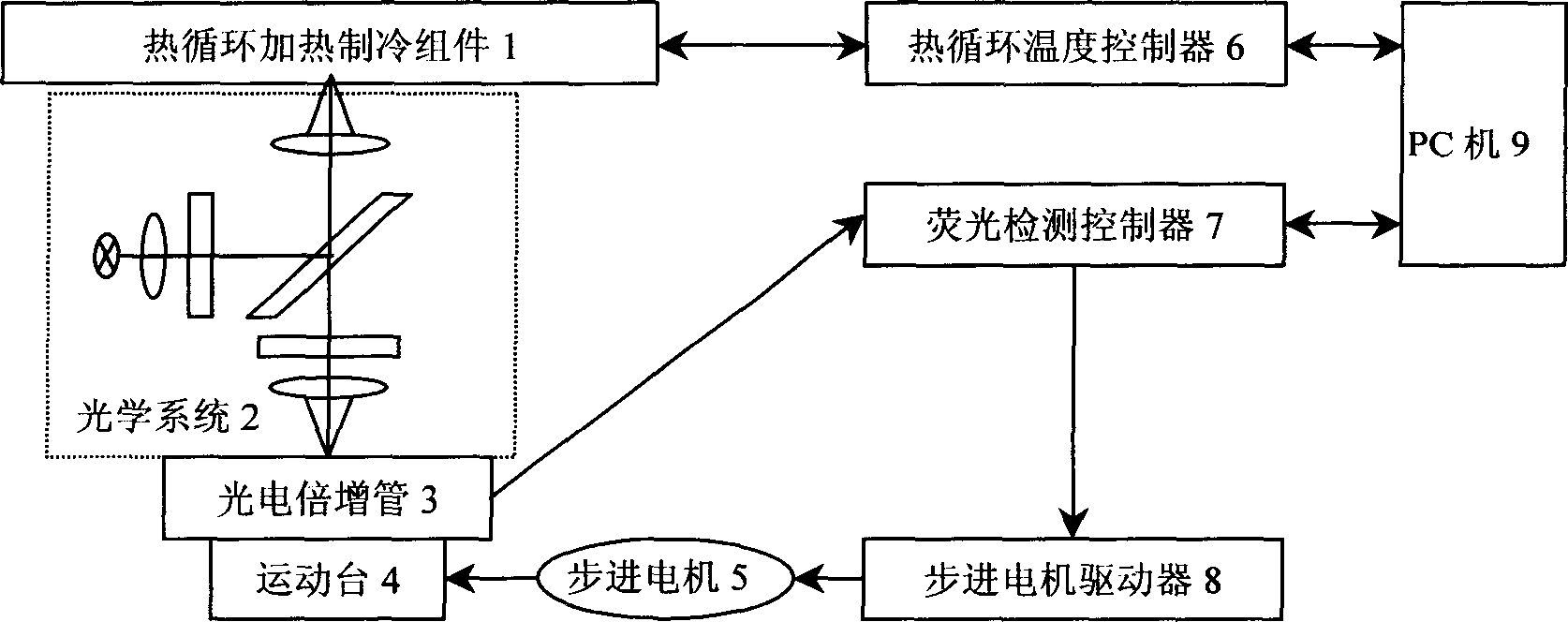
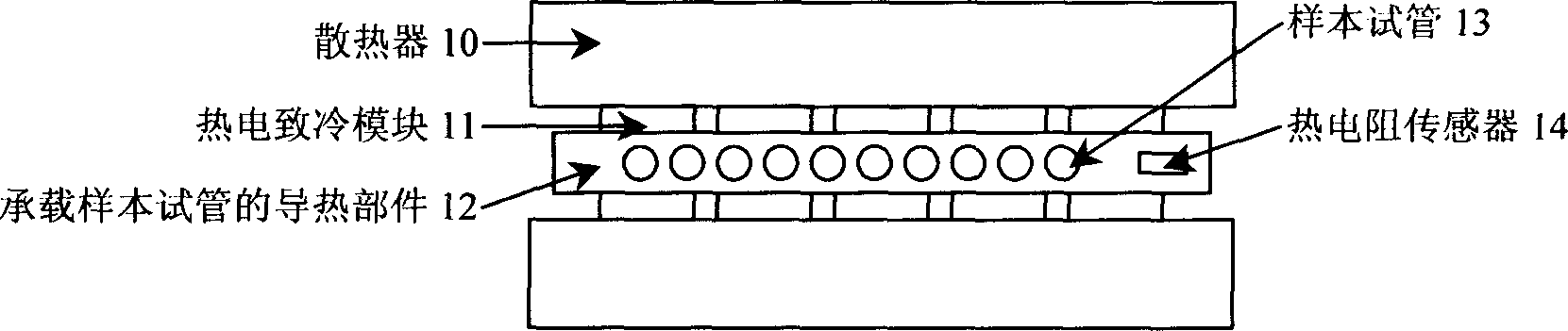
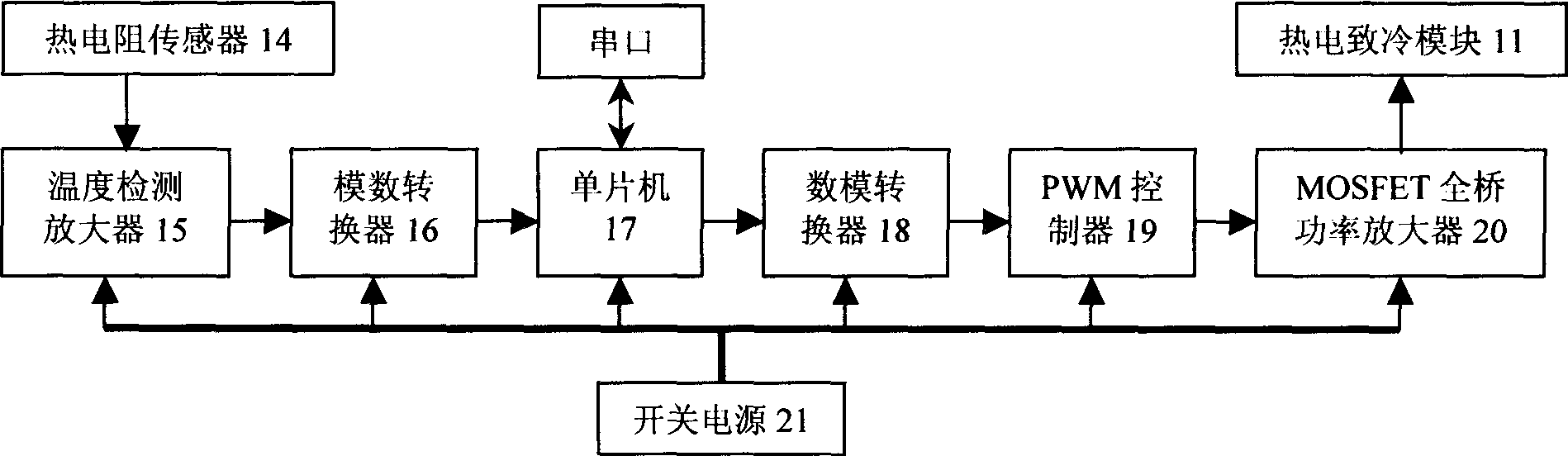
Heating cycle controlled polymerase chain reaction biological detection system
InactiveCN1800411ARich requirementsFlexible useMicrobiological testing/measurementFluorescence/phosphorescenceCycle controlFluorescence
The invention relates to a heat cycle control polyase chain reaction biological detection system which is formed by a heat cycle heating freezing component (1), an optical system (2), an electron-multiplier phototube (3), a moving table (4), a step motor (5), a heat cycle temperature controller (6), a fluorescence detecting controller (7), a step motor driver (8) and a PC machine (9). It adopts fluorescence real-time detecting mode to analyze the augmentation of PCR form; it adopts thermoelectric semiconductor cryogenic technique to achieve PCR course and uses high-fidelity photoelectric system to detect the fluorescence signal; it adopts micro machine as upper machine to do heat cycle craft controlling and adopts light intensity to real-time detect the PCR biology detecting system.
Owner:INST OF OPTICS & ELECTRONICS - CHINESE ACAD OF SCI
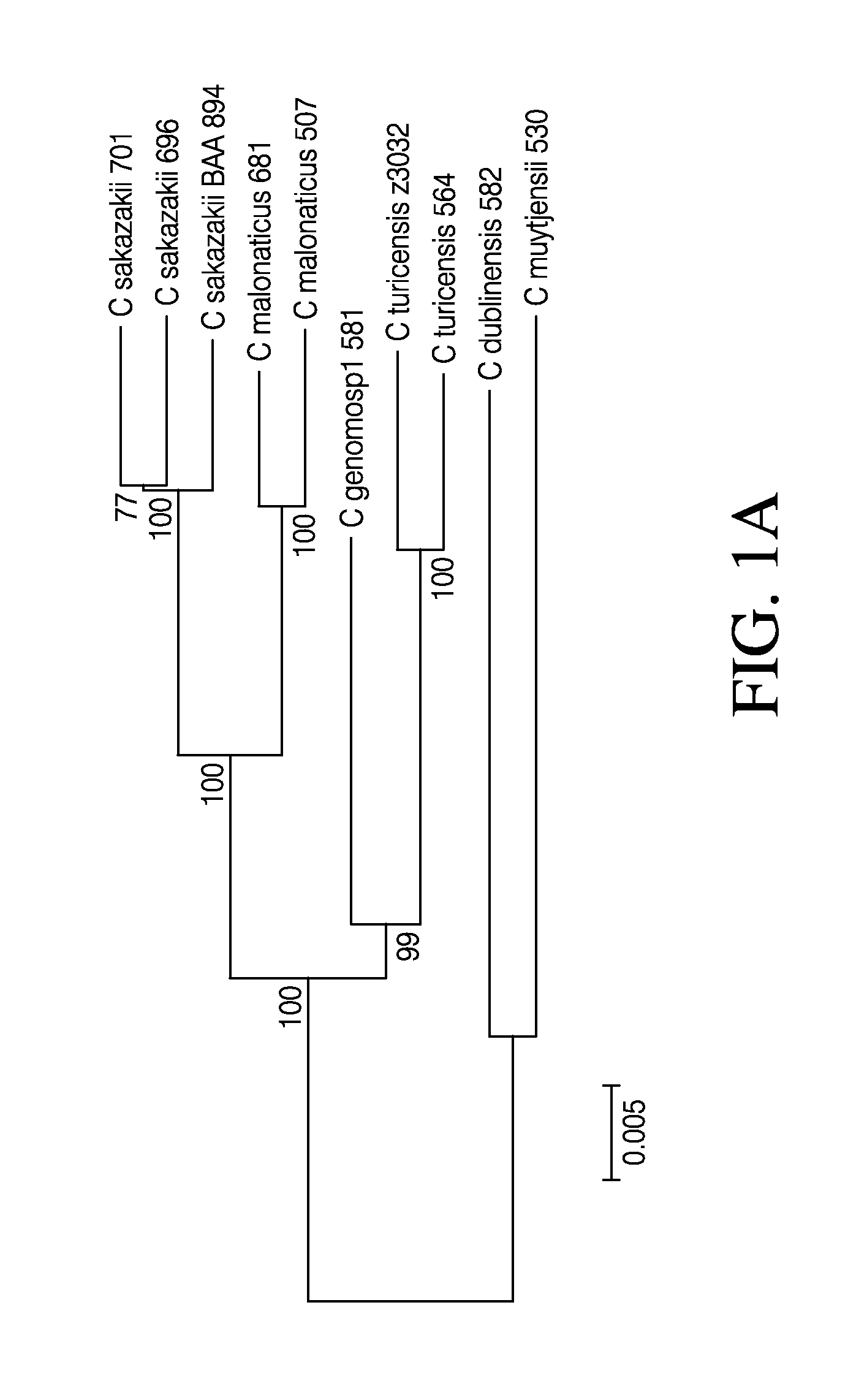
Compositions and methods for detection of cronobacter spp. and cronobacter species and strains
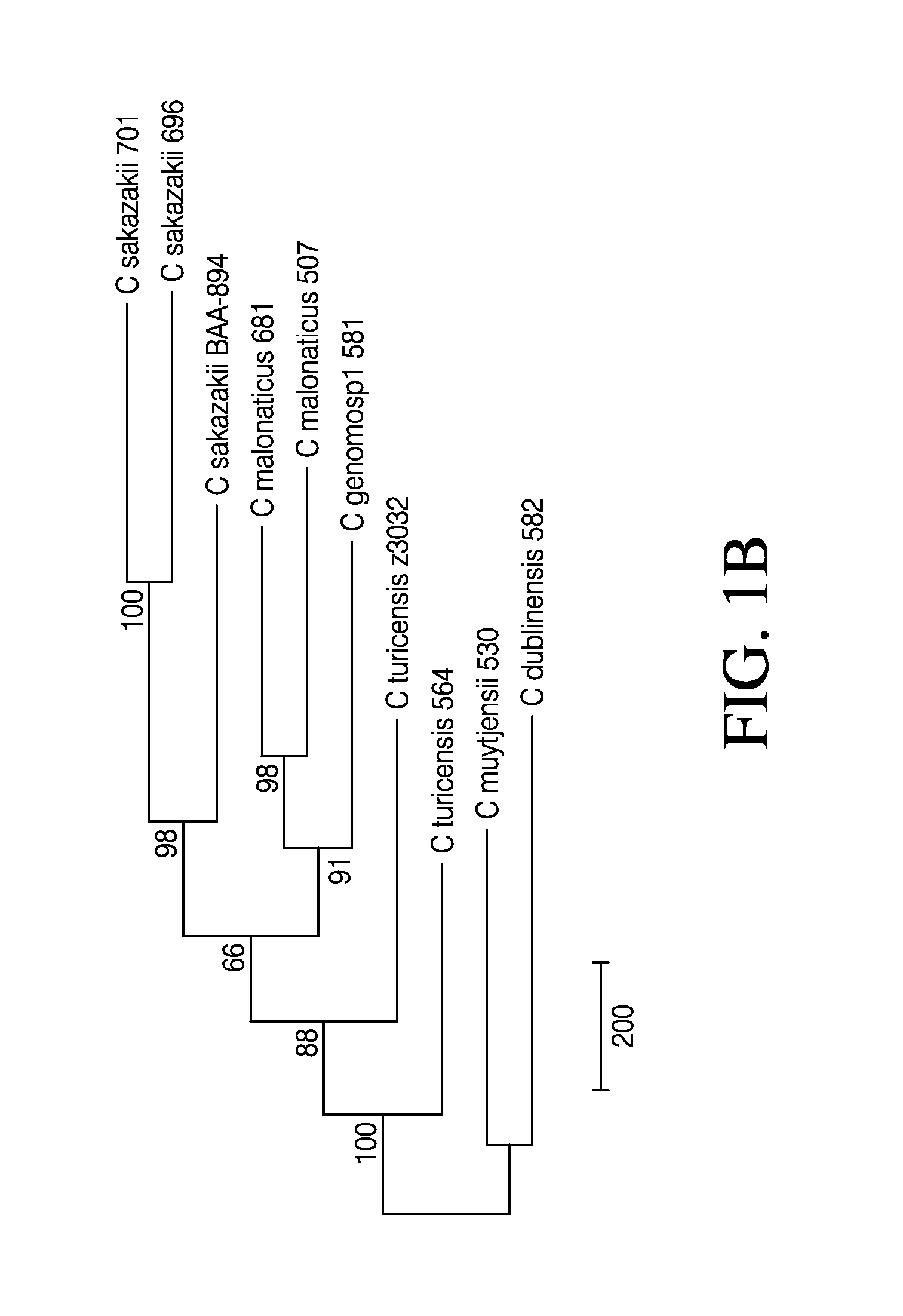
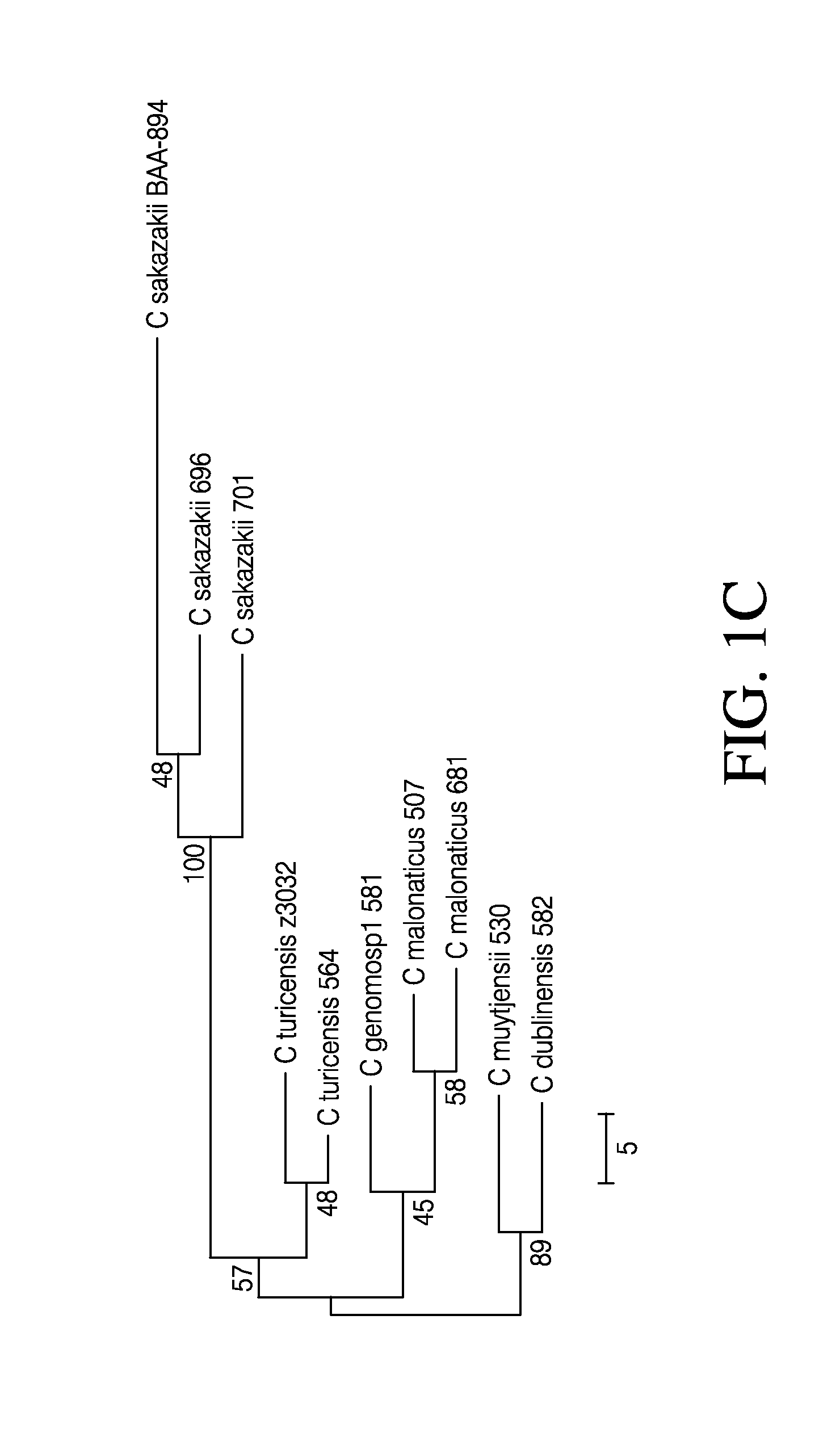
Disclosed are genomic sequences for nine strains of Cronobacter spp. (C. sakazakii—696, 701, 680; C. malonaticus—507, 681; C. turicensis—564; C. muytjensii—530; C. dublinensis—582; C. genomosp1—581) and compositions, methods, and kits for detecting, identifying and distinguishing Cronobacter spp. strains from each other and from non-Cronobacter spp. strains. Some embodiments describe isolated nucleic acid compositions unique to certain Cronobacter strains as well as compositions that are specific to all Cronobacter spp. Primer and probe compositions and methods of use of primers and probes are also provided. Kits for identification of Cronobacter spp. are also described. Some embodiments relate to computer software methods for setting a control based threshold for analysis of PCR data.
Owner:LIFE TECH CORP
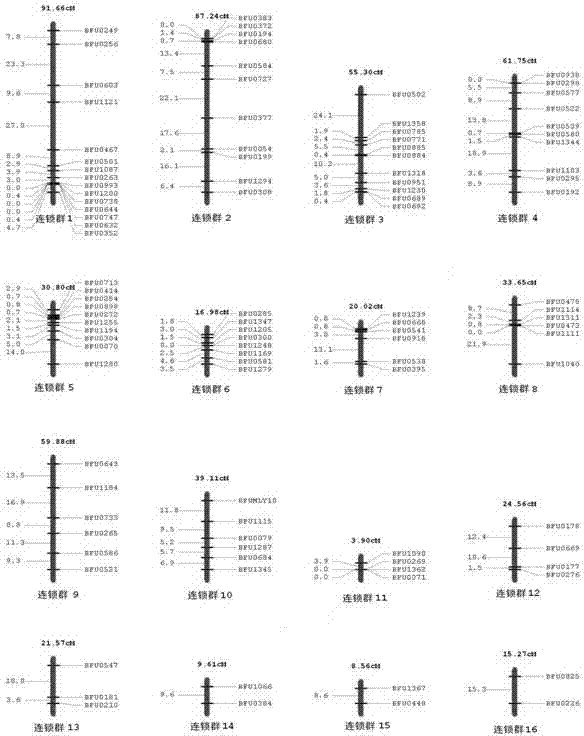
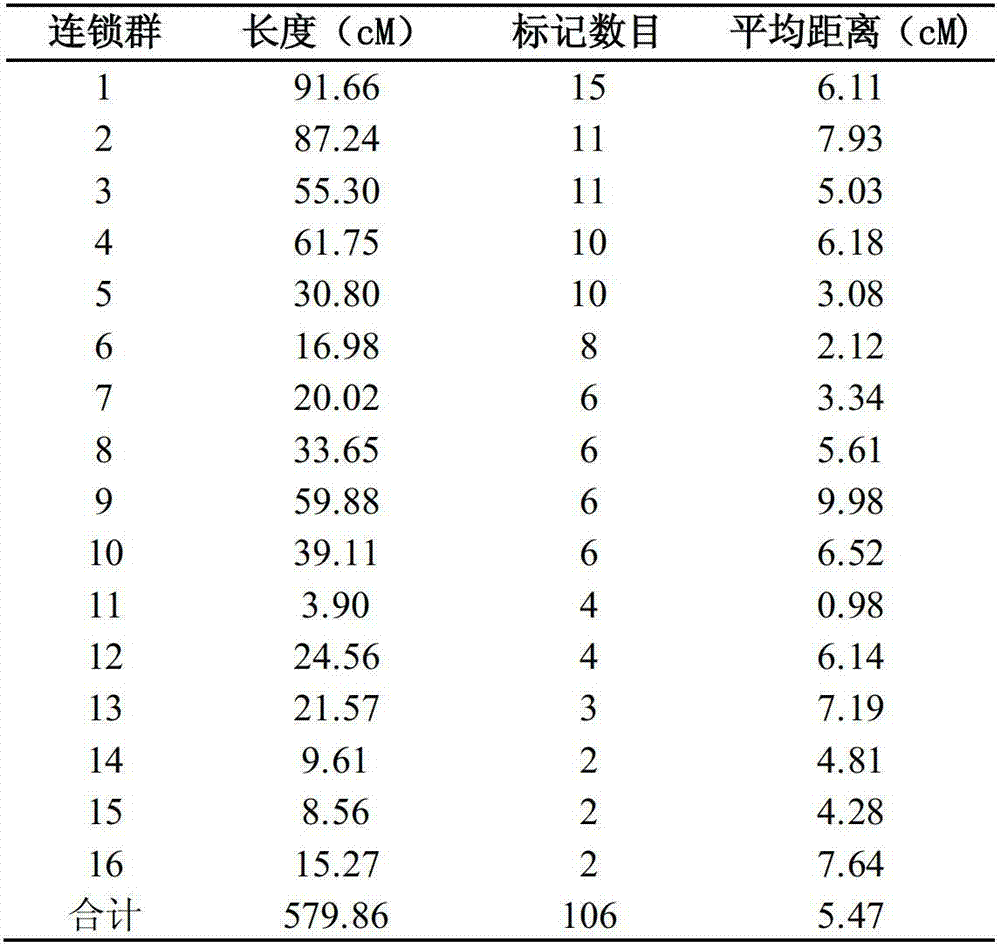
Construction method of date tree SSR (Simple Sequence repeat) marker molecular genetic map
InactiveCN103173562AImprove utilization efficiencyImprove experimental efficiencyMicrobiological testing/measurementGenetic diversityGenomic DNA
The invention discloses a construction method of a date tree SSR (Simple Sequence repeat) marker molecular genetic map. The construction method comprises the following steps of: (1) extracting genomic DNAs (deoxyribonucleic acids) of parents and offsprings of a date tree to be detected; (2) screening SSR primers by using parent materials; (3) carrying out PCR (Polymerase Chain Reaction) amplification by respectively using the SSR primers and respectively taking the genomic DNAs (deoxyribonucleic acids) of the parents and offsprings of the date tree to be detected as templates; and (4) analyzing PCR amplification results by using biology software, and constructing the date tree SSR marker molecular genetic map. According to the construction method, the high-efficiency low-cost date tree SSR marker technical system is constructed, and obtained SSR markers and a core primer group have the advantages of high efficiency, stability, co-dominance and the like. The set of markers can be used for researches such as the date tree genetic map construction, genetic diversity analysis, identification of variety, molecular mark assisted breeding and the like and promoting the development of date tree genetics and breeding technique.
Owner:BEIJING FORESTRY UNIVERSITY
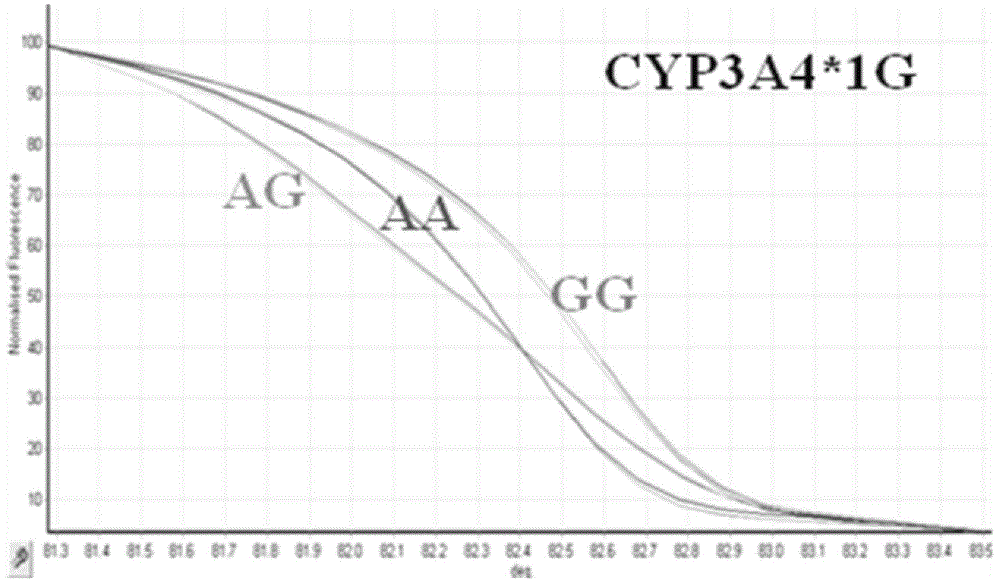
HRM method for detecting genetic polymorphism of CYP3A4*1G and MDR1C1236T
InactiveCN105274190AEasy and fast to provideMicrobiological testing/measurementUse medicationDissolution
The invention belongs to the field of molecular biology and medicine, and discloses an HRM analysis technology for detecting genetic polymorphism of CYP3A4*1G and MDR1C1236T. The method includes the following steps: (1) searching DNA sequences of target genes; (2) designing and screening appropriate primer pairs aiming at mutation loci of the target genes; (3) extracting sample DNA, and carrying out gene amplification by using the primers screened in the step (2); (4) analyzing whether a PCR product is obtained through specific amplification by an agarose gel electrophoresis method; and (5) according to a high-resolution dissolution curve method, detecting the mutation loci of the amplified target genes, and selecting an SYTO-9 saturated fluorescent dye. A sequence specific probe is not needed during testing, sequencing is not needed, the test is not limited by mutation base loci and types, genetic information is simply and quickly provided for clinical individualized medication, and a help is provided for rational drug use and clinical drug monitoring.
Owner:AFFILIATED HUSN HOSPITAL OF FUDAN UNIV
Method for detecting serotyping of streptococcus pneumoniae
ActiveCN103290115AMicrobiological testing/measurementMicroorganism based processesMultiplex ligation-dependent probe amplificationFluorescence
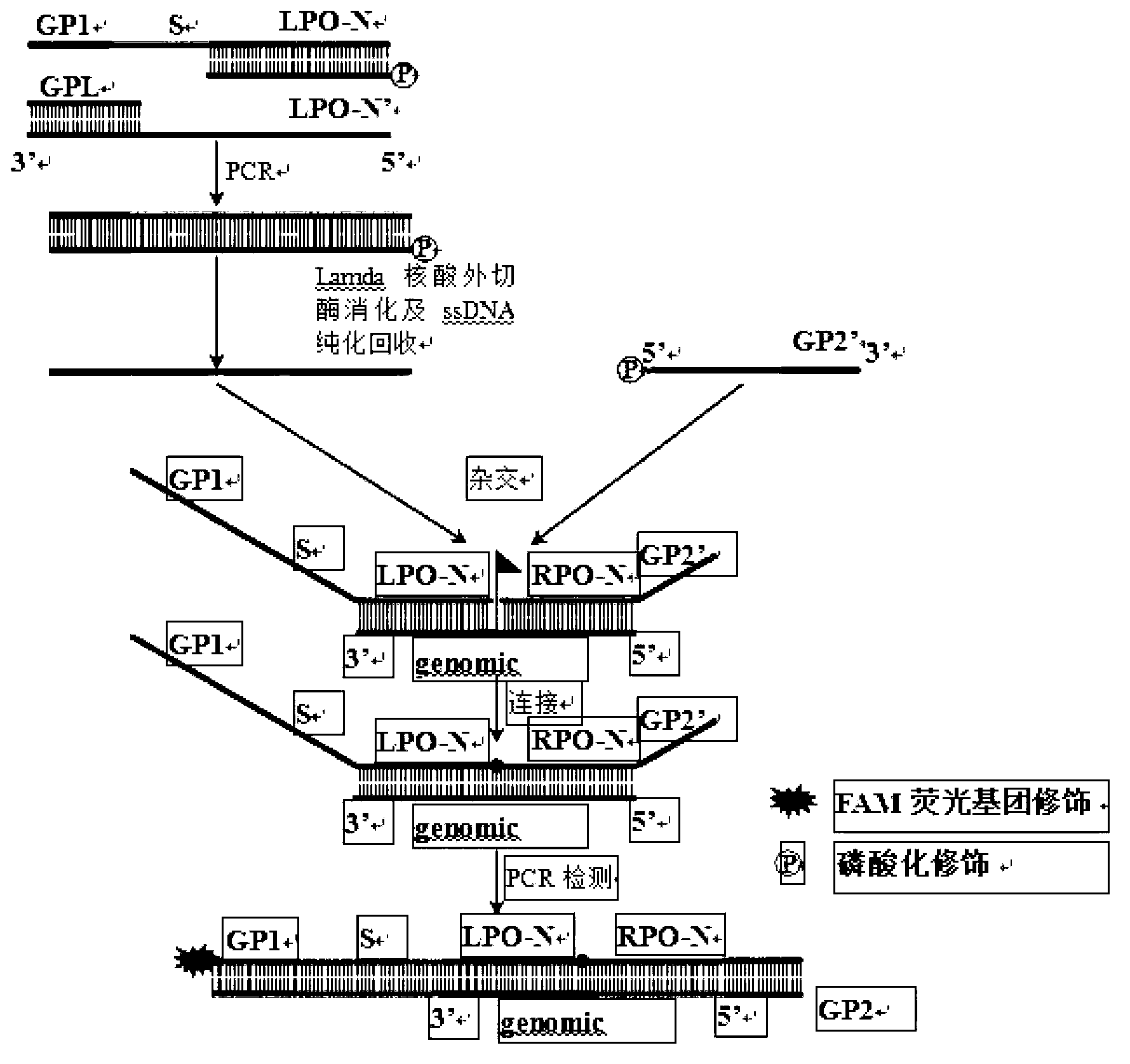
The invention provides a method for carrying out serotyping on streptococcus pneumoniae by using MLPA (multiplex ligation dependent probe amplification). The method comprises the following steps of: firstly, extracting a sample genome DNA of streptococcus pneumoniae to be detected; carrying out pre-amplification on the extracted genome DNA by using a PCR pre-amplification primer; carrying out hybridization on pre-amplification products of the genome DNA by using an MLPA probe; adding a corresponding fluorescence detecting probe in a reaction system; connecting hybridized MLPA probes by using ligase, and carrying out PCR amplification on the hybridized and connected MLPA probes by using an MLPA universal amplification primer; and analyzing PCR products by using a multicolor fluorescence dissolution curve, so that the serotyping on streptococcus pneumoniae can be realized. The detecting method disclosed by the invention is good in specificity and high in sensitivity; according to the method, 10 kinds of serotypes can be subjected to genotyping simultaneously just in 2-4 hours, so that multiple uncapping operations in traditional MLPA detection are avoided, and the pollution possibility is reduced, therefore, the method can meet high-throughput sample detection, and is especially applicable to inspection departments.
Owner:江西南兴医疗科技有限公司 +1
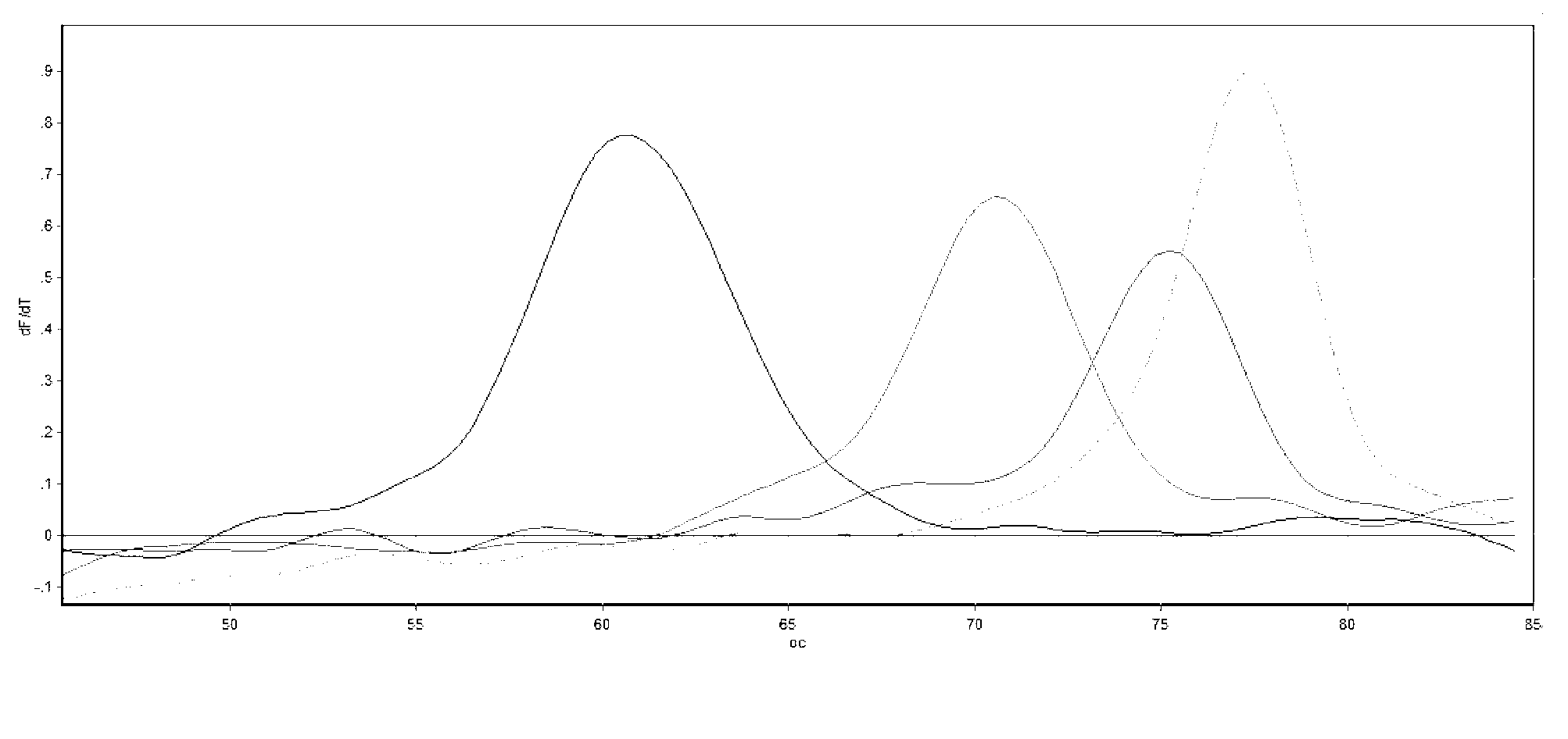
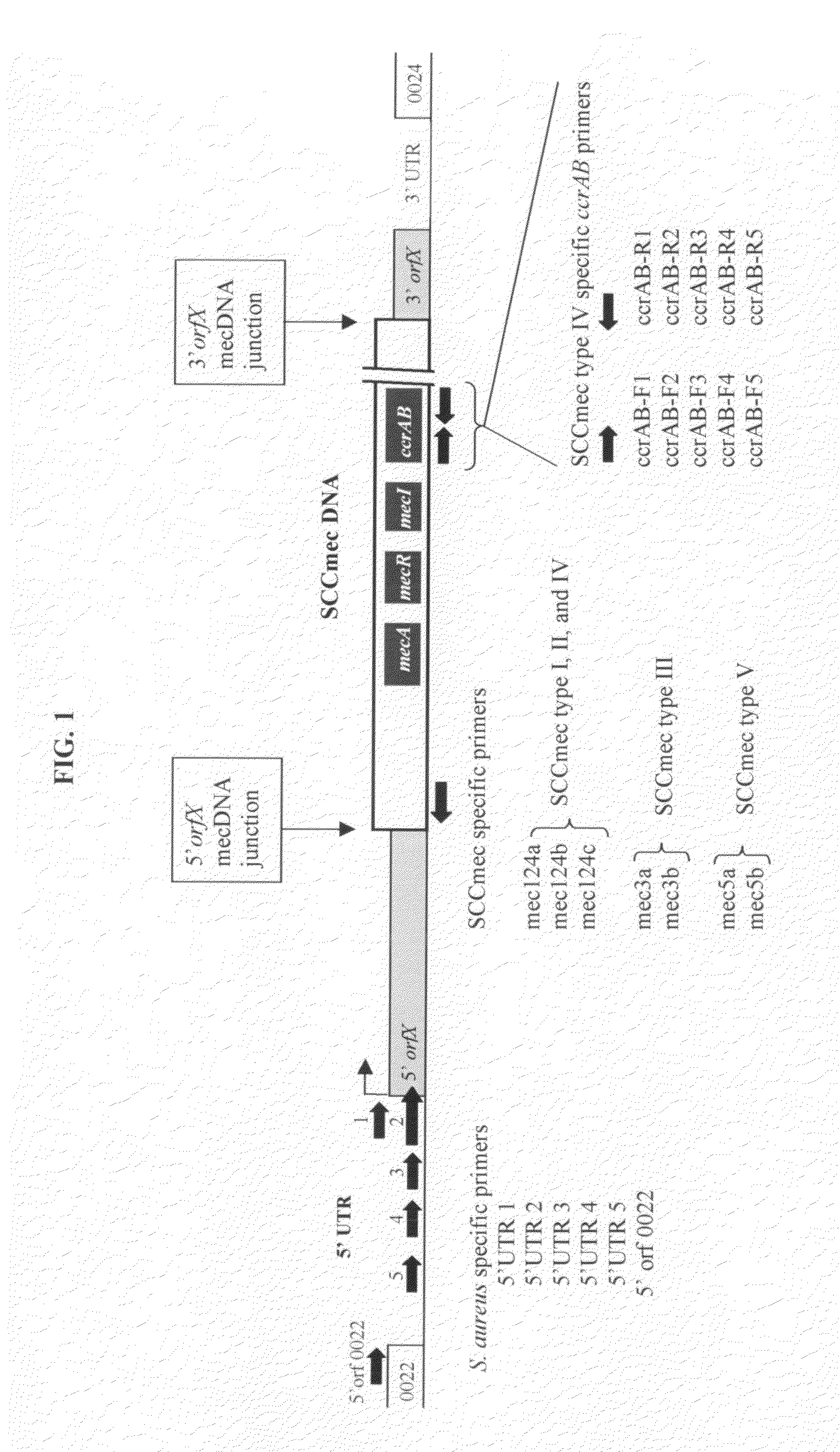
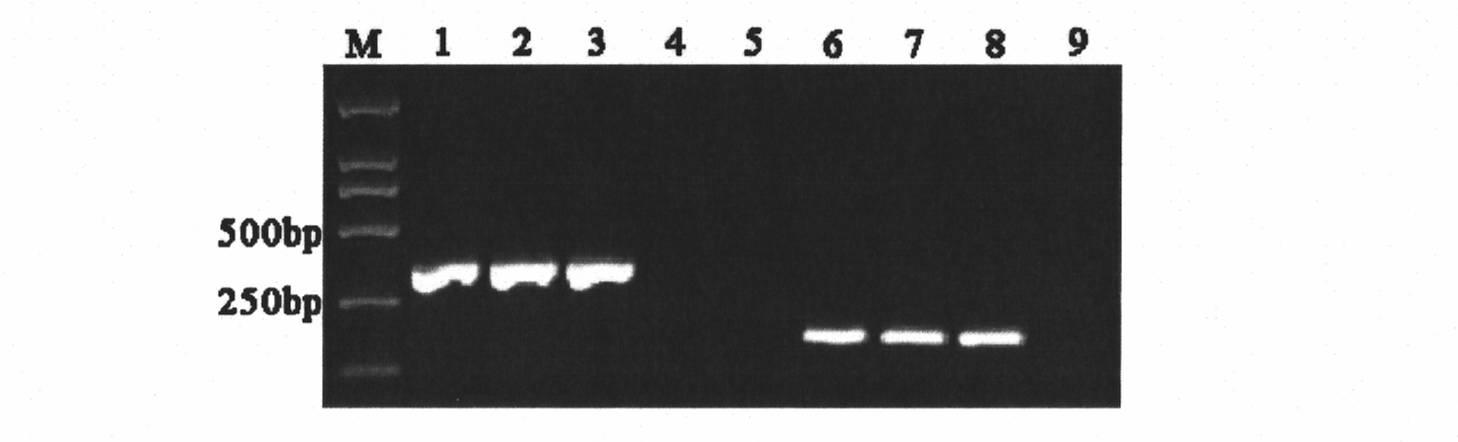
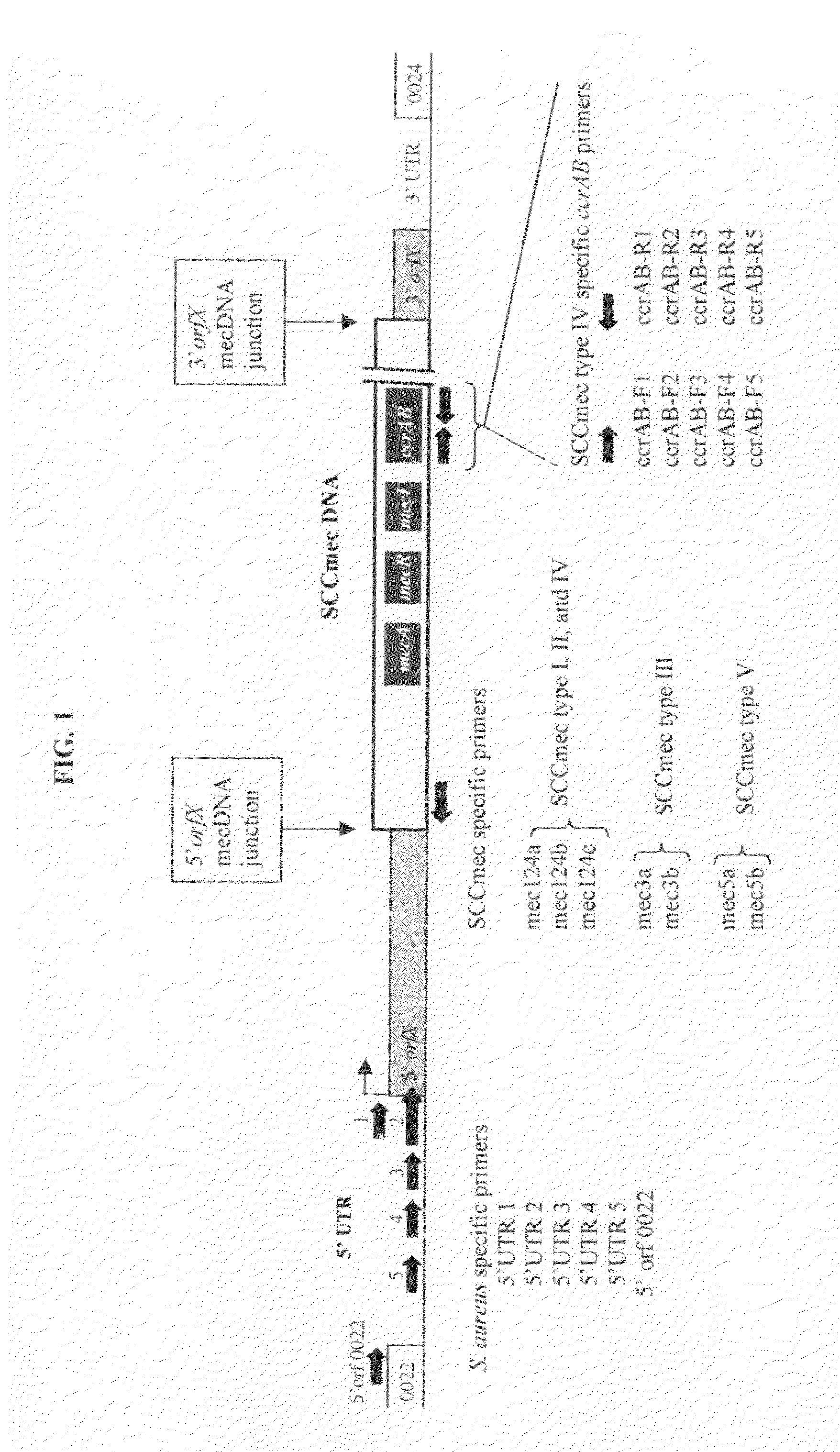
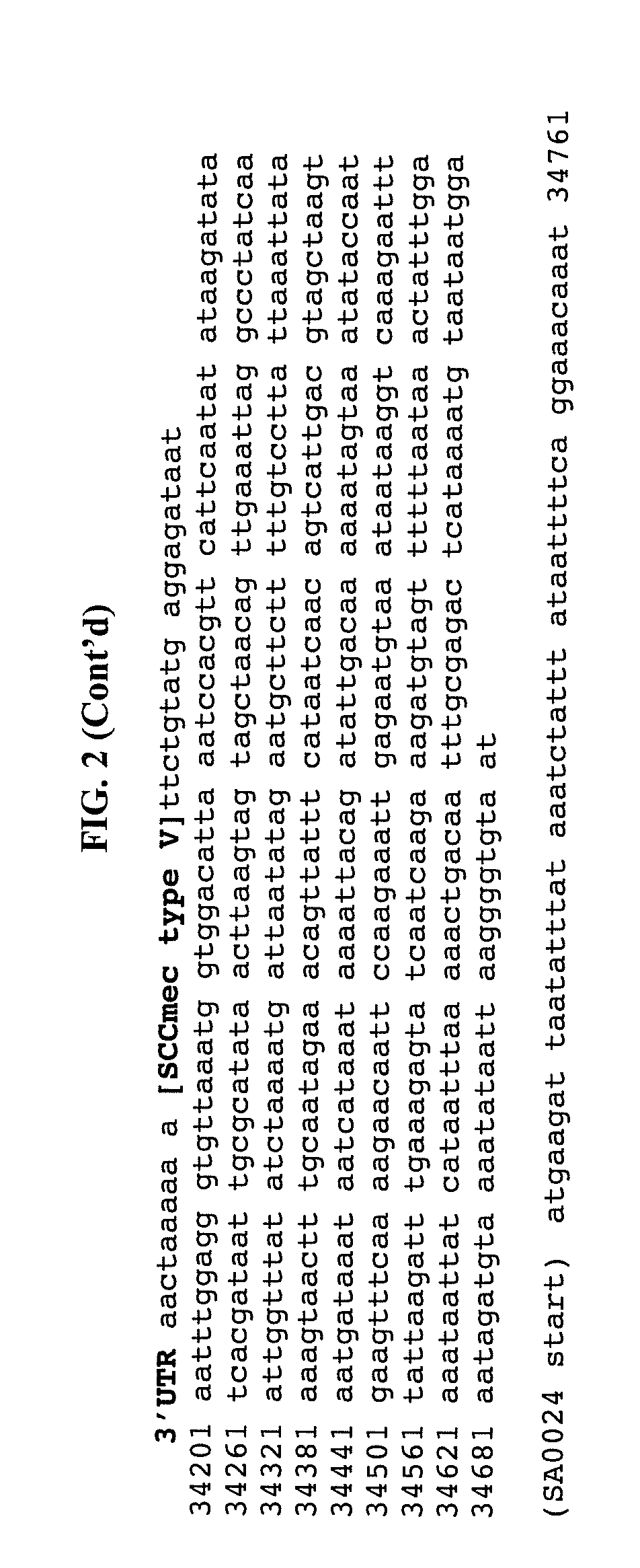
Method of determining types I, II, III, IV or V or methicillin-resistant staphylococcus aureus (MRSA) in a biological sample
ActiveUS20110312876A1Simple and sensitive screeningMuch sensitive and accurateAntibacterial agentsBiocidePresent methodStaphylococcus cohnii
Disclosed are diagnostic methods for determining a subtype of methicillin-resistant Staphylococcus aureus (MRSA) in a biological sample of a mammal. Methods include providing a biological sample of the mammal, performing a PCR analysis of the biological sample, and analyzing the PCR amplicons with respect to their sizes so as to determine for type I, type II, type III, type IV or type V MRSA that may be present in the biological sample. Further example embodiments include using at least one mecA primer pair and / or using at least one Staphylococcus aureus nuc primer pair in the PCR analysis. Further disclosed are methods for screening populations for MRSA, and methods of treating a mammal testing positive for Type IV MRSA. Also disclosed are kits for determining a MRSA subtype in a mammal and isolated primers that may be used in the present methods and kits.
Owner:MEDICAL DIAGNOSTIC LAB
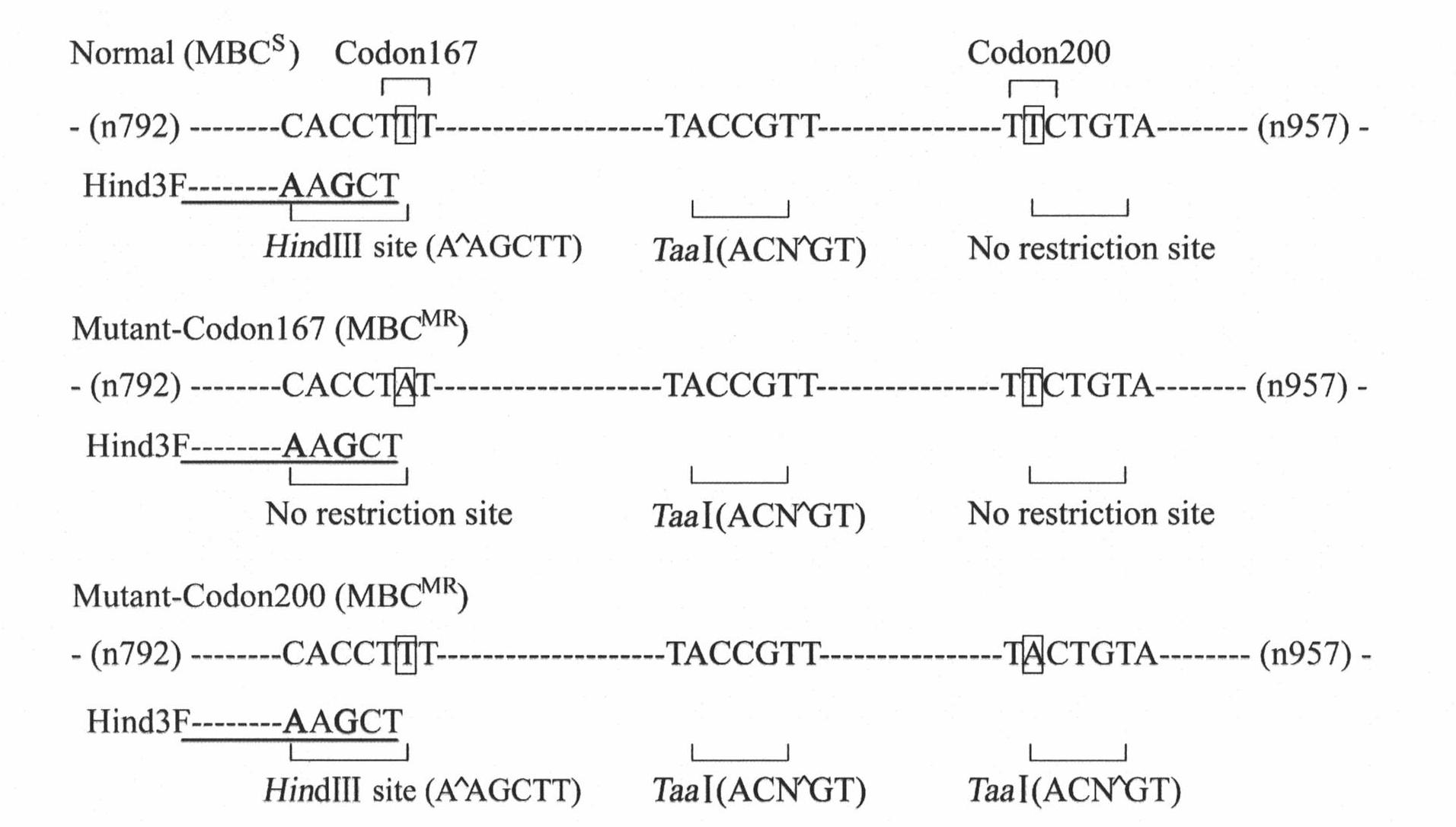
Molecule detection method of Fusarium graminearum to medium resistance level bacterial strain of carbendazim
InactiveCN101985653ATimely guidanceReasonable guidanceMicrobiological testing/measurementElectrophoresisBacterial strain
The invention belongs to a molecule detection method of Fusarium graminearum to medium resistance level bacterial strain of carbendazim. The method can be used for monitoring drug resistance of Fusarium graminearum to carbendazim and prevalence warning, wherein Fusarium graminearum can cause wheat scab. The detection method includes the following three main steps: (1) nuclear genome DNA of bacterial strain to be detected is respectively extracted: (2) internal and external primer pairs are utilized to carry out nested PCR, thus obtaining a target segment; (3) the target segment is respectively restricted by restriction enzymes HindIII and TaaI (Tsp4CI), and genotype of medium resistance level bacterial strain can be accurately identified by virtue of a restriction product electrophoresis spectrum. By adopting PIRA-PCR (primer-introduced restriction analysis PCR) technology, quantity of Fusarium graminearum with medium drug resistance in field and ratio thereof in group can be rapidly and accurately detected. Detection accuracy reaches more than 95%.
Owner:NANJING AGRICULTURAL UNIVERSITY
Method as well as molecular marker and primer for identifying rice male sterility type and three-line hybrid rice type
InactiveCN103436622AImprove identification efficiencyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural science3-deoxyribose
The invention discloses a method as well as a molecular marker and a primer for identifying a rice male sterility type and a three-line hybrid rice type, belonging to the field of molecular marker detection. Sequences of the molecular marker and the primer for identifying the rice cytoplasmic male sterility type and the three-line hybrid rice type are shown as SEQ ID NO.1-36. The identification method disclosed by the invention comprises the steps of extracting the total DNA (Deoxyribose Nucleic Acid) of a rice material; by taking the extracted total DNA as a template, using the primer for PCR (Polymerase Chain Reaction) amplification; analyzing PCR amplification results, if amplification banding patterns of two materials are completely consistent, the two materials have the same cytoplasm karyotype, namely have the same female parent source, otherwise, the two materials have different cytoplasm karyotypes, namely have different female parent sources. The method as well as the molecular marker and the primer for identifying the rice male sterility type and the three-line hybrid rice type are easy and convenient to operate, stable and reliable, can be used for identifying cytoplasmic male sterile lines and three-line hybrid rice varieties, have the characteristics of accuracy, high efficiency and rapidness, and thus improve the identification efficiency of the sterile lines and new varieties.
Owner:WUHAN UNIV
Polymerase chain reaction detection method of hepatitis B virus genotyping
ActiveCN102140537ASimple and fast operationHigh compliance rateMicrobiological testing/measurementFluorescence/phosphorescenceSerum samplesFluorescence
The invention provides a polymerase chain reaction (PCR) detection method of hepatitis B virus (HBV) genotyping. The method comprises the following steps: a primer is designed according to the entire genome sequence of the HBV genotype; in the existence of the primer, DNAs are extracted from a serum sample to perform PCR amplification in a fluorescence PCR meter, the PCR product is analyzed according to the melting curve, the genotype is judged according to the Tm value of the PCR product and the HBV genotyping can be realized. The PCR melting curve method used in the invention is convenient to operate, the detection result of the method has higher coincidence rate with the detection result obtained by adopting a commercial genotyping kit; and the method can be used in the clinical detection of the HBV genotype. On the basis of the PCR technology, the PCR melting curve method which is to judge the HBV genotype according to the Tm value, is provided in the PCR detection method, thus the HBV genotype can be identified conveniently, rapidly and accurately.
Owner:THE FIRST AFFILIATED HOSPITAL OF FUJIAN MEDICAL UNIV
Detection method and kit for BUD13-ZNF259 gene mononucleotide polymorphism and application thereof
InactiveCN104673882ASimple and fast operationImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationDiseaseFluorescence
The invention relates to a method for detecting BUD13-ZNF259 gene rs964184 mononucleotide polymorphism by a Tm-shift process. The method comprises the following steps: respectively adding GS-rich sequence to the 5' end of a specific primer, and pairing base of the 3' terminal with allele mutant of SNP (single-nucleotide polymorphism); and in the presence of the primer, carrying out PCR (polymerase chain reaction) amplification on DNA (deoxyribonucleic acid) extracted from the serum sample in a fluorescence PCR instrument, analyzing the PCR product by using a melting curve, and judging the genotype according to the Tm value of the PCR product. The primer can be used for preparing a kit for detecting susceptibility of coronary heart disease. The Tm-shift process is simple to operate, has the advantages of high accuracy, favorable repetitiveness and low cost, and can be used for promoting early prevention of diseases and providing references for drug therapy.
Owner:NINGBO UNIV
Method for fast detecting genome DNA residues in total RNA sample
ActiveCN108754010AImprove accuracyDetection is simple and fastMicrobiological testing/measurementDNA/RNA fragmentationInteinTotal rna
The invention provides a method for fast detecting genome DNA residues in a total RNA sample. The method comprises the steps that 1, according to genetic information of a to-be-detected biological material, a gene containing one or more intron sequence is selected as a target, and for the intron sequence, PCR primers are designed; 2, the total RNA of the to-be-detected biological material is extracted, the total RNA or cDNA obtained through reverse transcription of the total RNA is used as a template, and the primers in the step 1 are used for PCR amplification; 3, a PCR amplification productis analyzed. According to the method, only one pair of the primers are designed, so that whether or not the genome DNA residues exist in the total RNA and cDNA samples of the biological material can be detected simply, conveniently and fast; the method provides support for the accuracy of gene expression analysis and has important scientific significance for promoting fundamental research of the molecular biology.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
Method for identifying varieties of low-phenol cotton
InactiveCN105087803AImprove use valueMicrobiological testing/measurementAgricultural scienceTest material
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
Primer set for detecting bovine norovirus and bovine coronavirus and double RT-PCR method
ActiveCN108913815AStrong specificityIncreased sensitivityMicrobiological testing/measurementAgainst vector-borne diseasesAgricultural scienceNucleotide
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY MEDICINE HENAN ACAD OF AGRI SCI
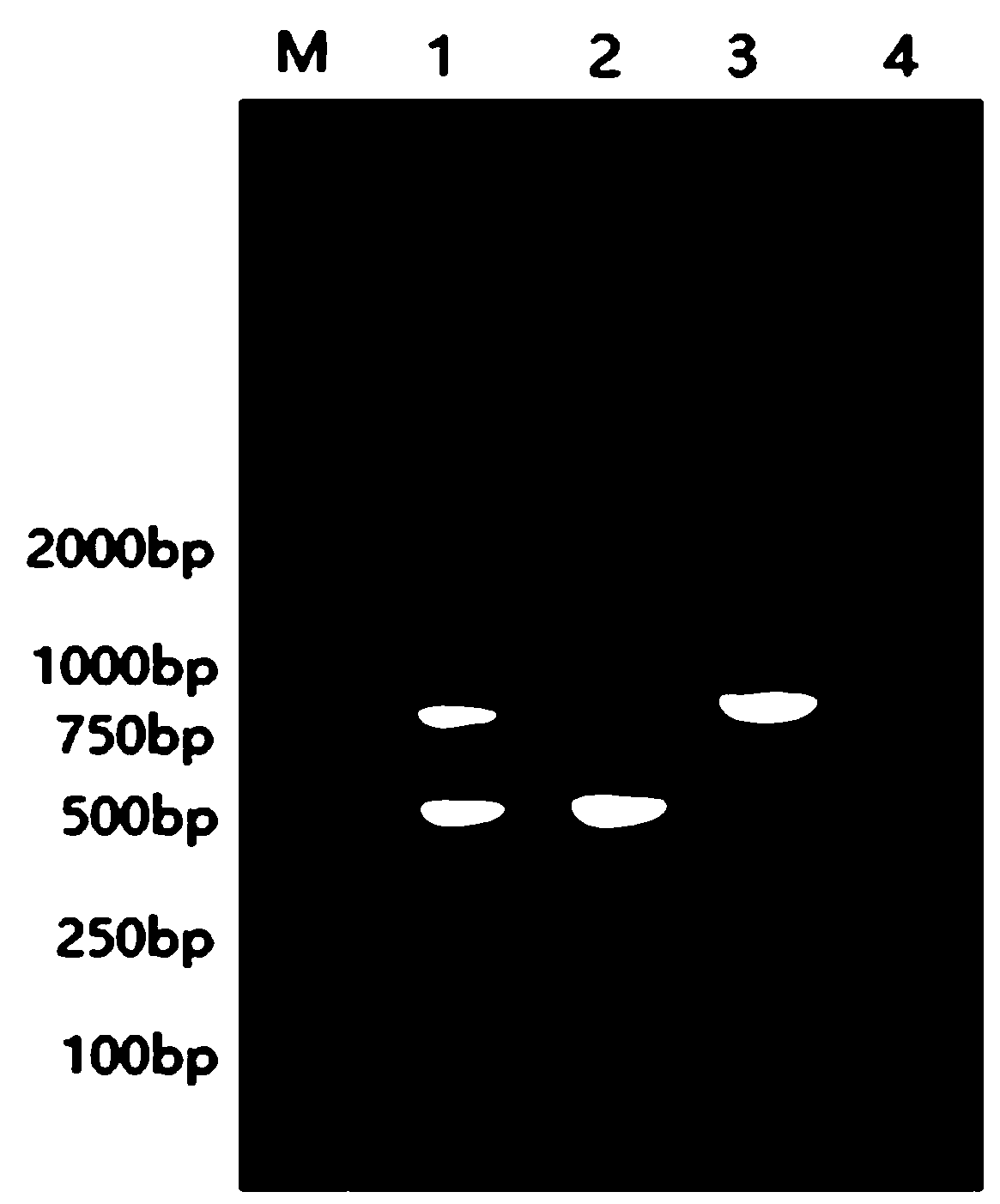
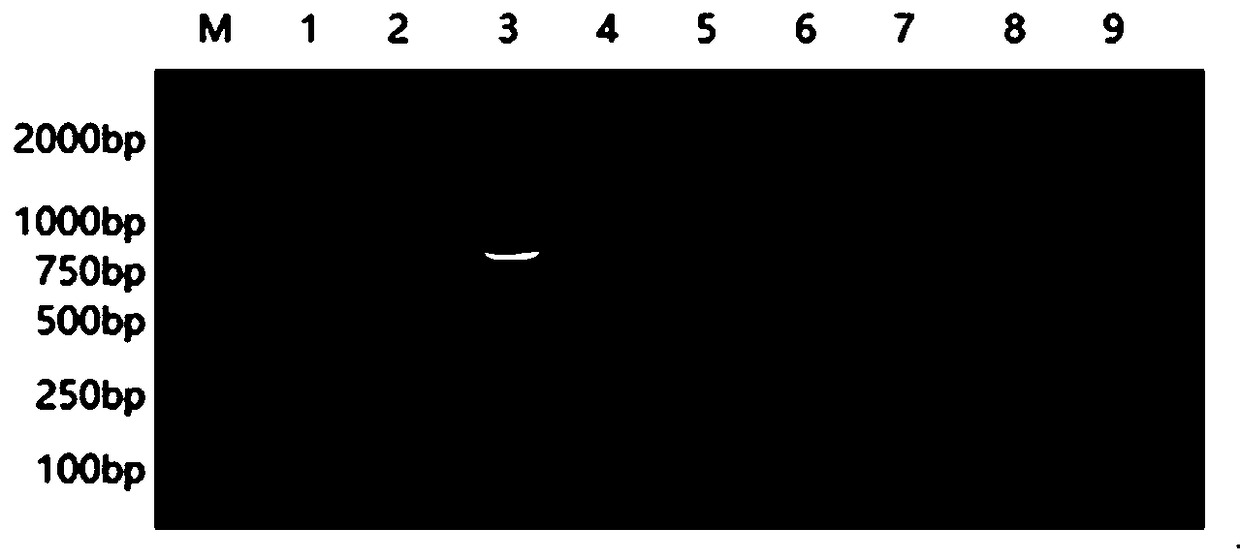
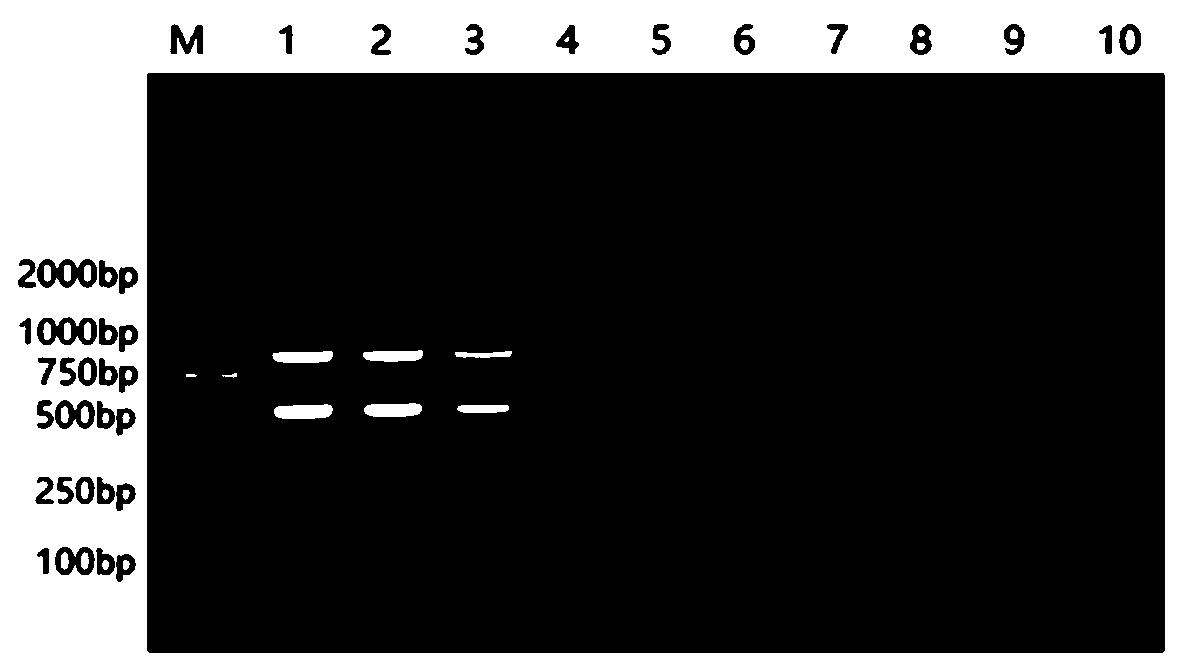
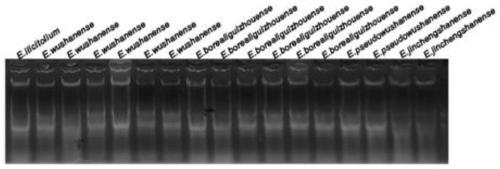
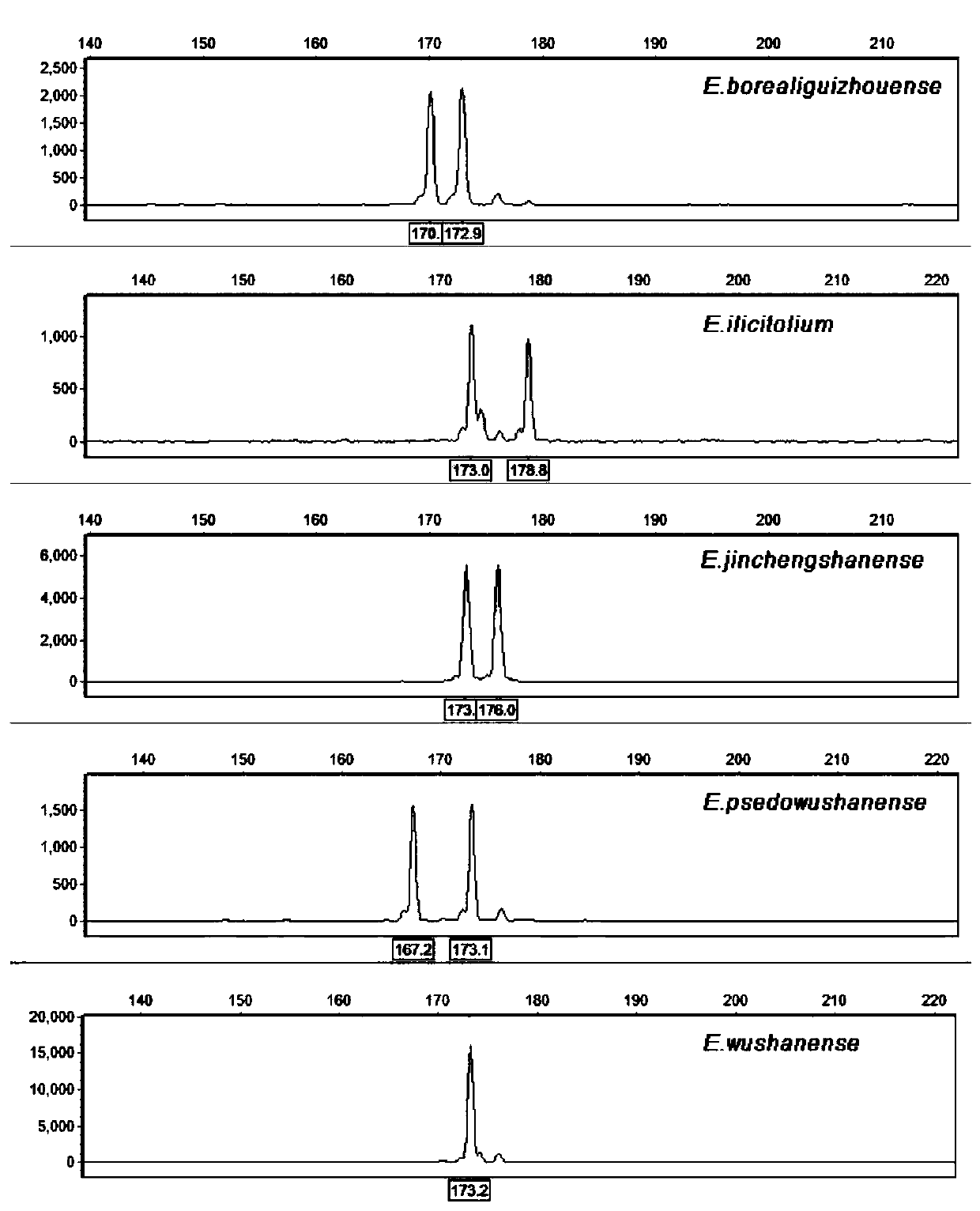
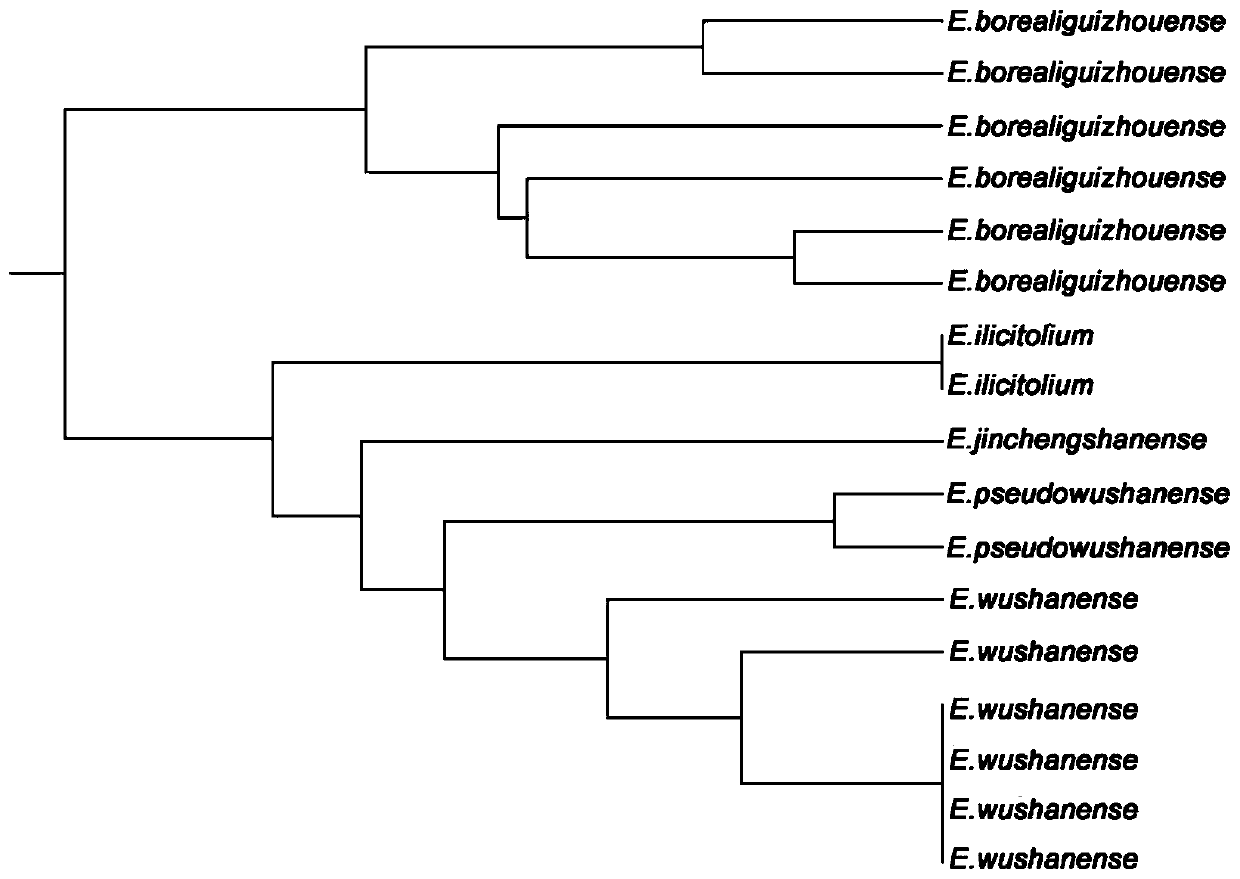
EST-SSR molecular identification method for E.wushanense and easily mixed species thereof
ActiveCN110205397AGood polymorphismStrong specificityMicrobiological testing/measurementAgainst vector-borne diseasesMolecular identificationPhylogenetic tree
The invention discloses an EST-SSR molecular identification method for E.wushanense and an easily mixed species thereof, and belongs to molecular markers and medicinal material identification in the technical field of molecular biology. An EST-SSR primer group for identification of E.wushanense and the easily mixed species thereof include 10 pairs of EST-SSR core primers and common M13 primers having the sequences of SEQ ID NO:1, 2...20 and 21. The identification method includes the steps: (1) extracting total DNA of a sample by a CTAB method; (2) amplifying by PCR by using total DNA of the sample to be determined as a template; (3) analyzing a product of PCR amplification; and (4) constructing a phylogenetic tree. An application comprises identification of E.wushanense and the easily mixed species thereof. The 10 SSR molecular markers provided by the invention have good polymorphism, high specificity and stable amplification results, can rapidly and accurately identify E.wushanense and the easily mixed species thereof, provide technical support for promotion of the quality standard of epimedium medicinal materials, and are beneficial to development and utilization of the epimediummedicinal materials.
Owner:WUHAN BOTANICAL GARDEN CHINESE ACAD OF SCI
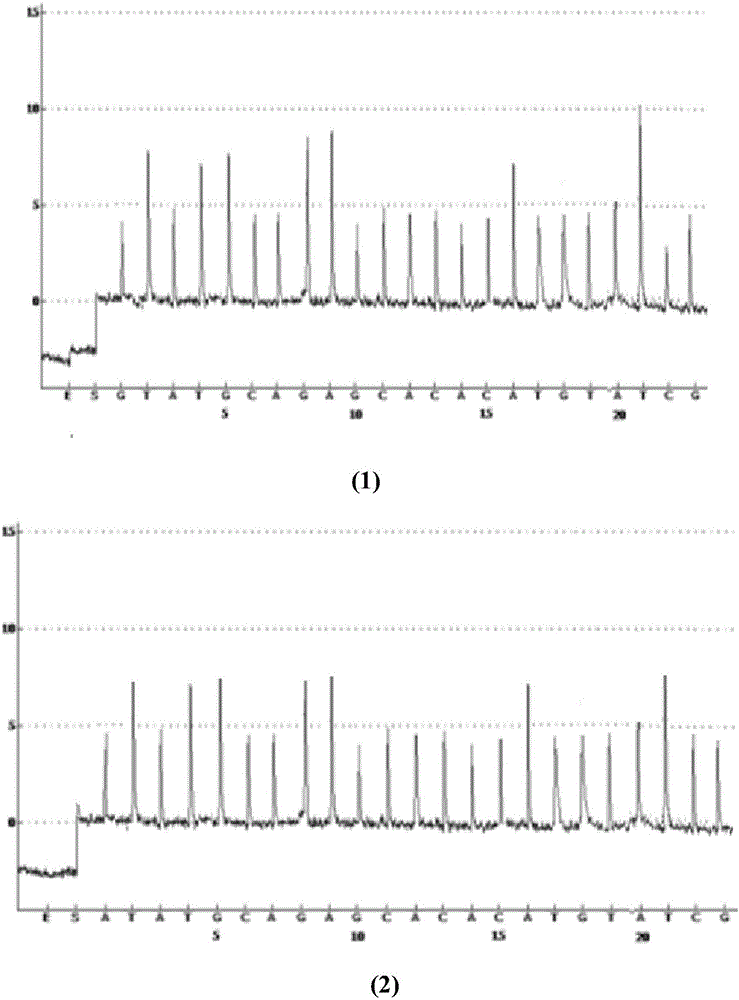
Method for analyzing haplotype of PCR product employing non-synchronous synthesis sequencing of two nucleotides
ActiveCN104762406ABroaden the scope of analysisEasy to operateMicrobiological testing/measurementNucleotideHaplotype
The invention discloses a method for analyzing haplotype of a PCR product employing non-synchronous synthesis sequencing of two nucleotides. Through real-time synthesis sequencing of two nucleotides, two different nucleotides are simultaneously added for each sequencing reaction; the synthesis sequencing of different DNA templates in the PCR product is not synchronous, so that information of different base groups on two adjacent SNP sites is provided by different sequencing reactions; and association analysis is carried out according to sequencing information on different SNP sites obtained by these non-synchronous synthesis sequencing reactions, so as to determine the haplotype of the PCR product. The analysis range is expanded; the operation is simple; the analysis cost can be reduced; the method is suitable for haplotype analysis of a plurality of mixing samples; the ratio of individual haplotypes in the mixing samples can be directly determined; and ucleic acid markers can be searched and screened from large samples.
Owner:SOUTHEAST UNIV
Analytical method for structures of microbial communities in composite filler in tower-type earthworm ecological filter
InactiveCN102559857AIdentify community changesGood removal effectMicrobiological testing/measurementMaterial analysis by electric/magnetic meansSequence analysisGel electrophoresis
The invention relates to an analytical method for compositions of microbial communities in composite fillers (soil, gravels and broken bluestones) in a tower-type earthworm ecological filter. The analytical method can be used for better reflecting the diversity and dynamic variation of microbial populations and specifically comprises the following steps of: (1). respectively taking samples in different filler layers of a tower-type earthworm ecological filter for preprocessing; (2) extracting the total DNAs (Deoxyribose Nucleic Acids) of genomes by using a MOBIO DNA extraction kit; (3) carrying out nest type PCR (Polymerase Chain Reaction) amplification on the DNAs of the samples by using a design primer of a 16S RNA V3 region; (4) analyzing PCR amplification products by using a denaturing gradient gel electrophoresis (DGGE) technology, and analyzing an obtained DGGE atlas by using Quantity One software; (5) cutting and recycling characteristic DGGE bands, carrying out sequencing analysis on the DNAs of the samples after carrying out PCR amplification again, and judging the variety of microorganisms.
Owner:郭飞宏
Specific SS-COI (species-specific-COI) primer based PCR rapid detection method for Frankliniella intonsa
InactiveCN107663547ARapid identificationIdentify food chain relationshipsMicrobiological testing/measurementDNA/RNA fragmentationSpecific detectionFrankliniella intonsa
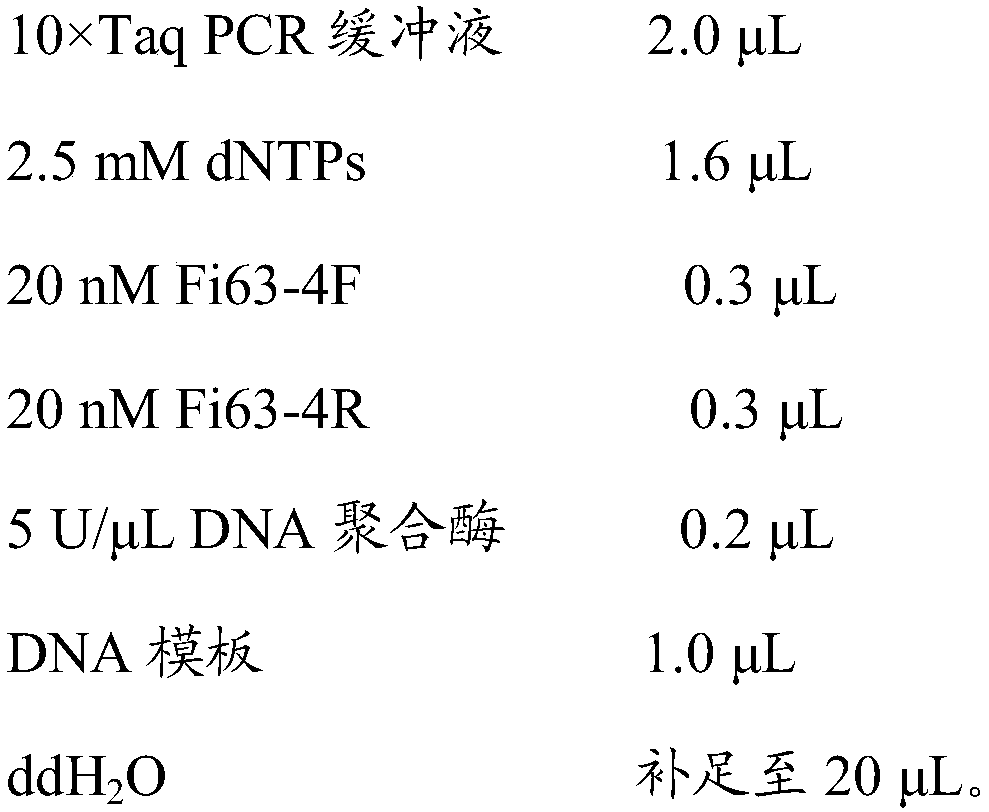
The invention provides specific SS-COI (species-specific-COI) primers for Frankliniella intonsa, a kit containing the primers and used for specifically detecting Frankliniella intonsa, an applicationof the primers or the kit containing the primers to specific detection of the Frankliniella intonsa and a specific SS-COI primer based PCR rapid detection method for the Frankliniella intonsa. The SS-COI primers comprise Fi63-4F:5'-CGCTAAGTATTTTAATTCGG-3'(SEQID No.1) and Fi63-4R:5'-AGTGGCACTAGTCAGTTTCC-3'(SEQ ID No.2). The specific SS-COI primer based PCR rapid detection method comprises the following steps: 1) extracting DNA of a sample; 2) performing a PCR amplification reaction with DNA extracted in the step 1) as a template as well as the primers; 3) analyzing a PCR product.
Owner:INST OF PLANT PROTECTION OF XINJIANG ACADEMY OF AGRI SCI
A kind of detection method of Streptococcus pneumoniae serotype
ActiveCN103290115BMicrobiological testing/measurementMicroorganism based processesMultiplex ligation-dependent probe amplificationFluorescence
The invention provides a method for carrying out serotyping on streptococcus pneumoniae by using MLPA (multiplex ligation dependent probe amplification). The method comprises the following steps of: firstly, extracting a sample genome DNA of streptococcus pneumoniae to be detected; carrying out pre-amplification on the extracted genome DNA by using a PCR pre-amplification primer; carrying out hybridization on pre-amplification products of the genome DNA by using an MLPA probe; adding a corresponding fluorescence detecting probe in a reaction system; connecting hybridized MLPA probes by using ligase, and carrying out PCR amplification on the hybridized and connected MLPA probes by using an MLPA universal amplification primer; and analyzing PCR products by using a multicolor fluorescence dissolution curve, so that the serotyping on streptococcus pneumoniae can be realized. The detecting method disclosed by the invention is good in specificity and high in sensitivity; according to the method, 10 kinds of serotypes can be subjected to genotyping simultaneously just in 2-4 hours, so that multiple uncapping operations in traditional MLPA detection are avoided, and the pollution possibility is reduced, therefore, the method can meet high-throughput sample detection, and is especially applicable to inspection departments.
Owner:江西南兴医疗科技有限公司 +1
Multiple PCR method for detecting quinolone antibiotics campylobacter jejuni and kit
ActiveCN103525930BAccurate detectionHigh sensitivityMicrobiological testing/measurementAgainst vector-borne diseasesNucleotideAntibiotic Y
The invention integrates mispairing amplification mutation analysis PCR (MAMA-PCR) and a multiple PCR technique, and provides a method for rapidly detecting quinolone antibiotics campylobacter jejuni caused by point mutation of nucleotide and a kit. The kit comprises a primer composition with the nucleotide sequence shown by SEQ ID No.1-5 and SEQ ID No.7 in the sequence list and a quality control group primer composition with the nucleotide sequence shown by SEQ ID No.1-6 in the sequence list. By adopting the detection method and the kit, campylobacter jejuni of the quinolone antibiotics can be rapidly identified from suspected bacteria, and complex procedures of the traditional detection method using strain identification and drug sensitive test as the representative are simplified. The method and the kit have favorable specificity and sensitivity, and are very suitable for rapidly detecting suspected bacteria at the frontline to provide accurate medicine instructions.
Owner:CHINA AGRI UNIV
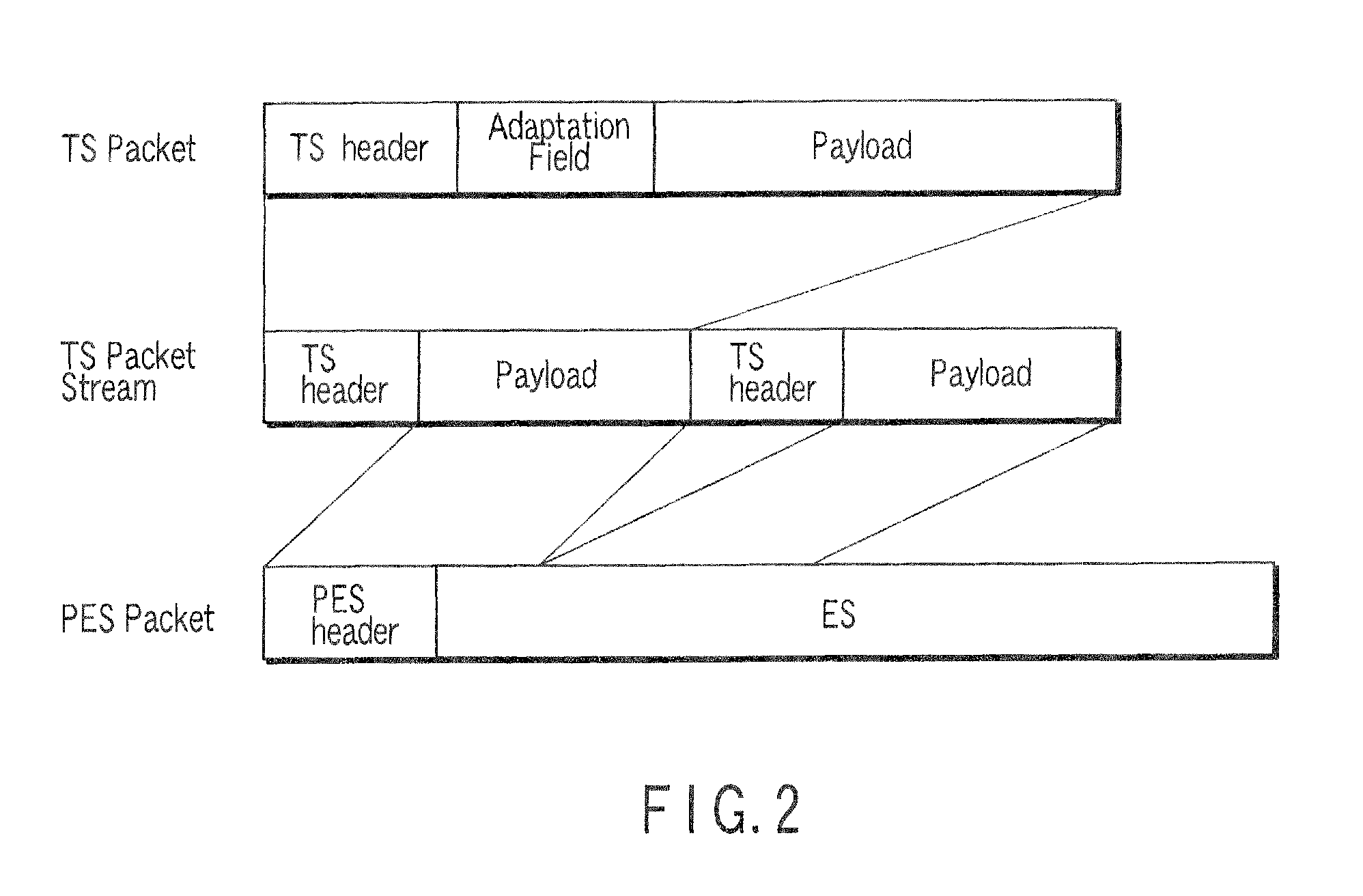
Stream recording apparatus
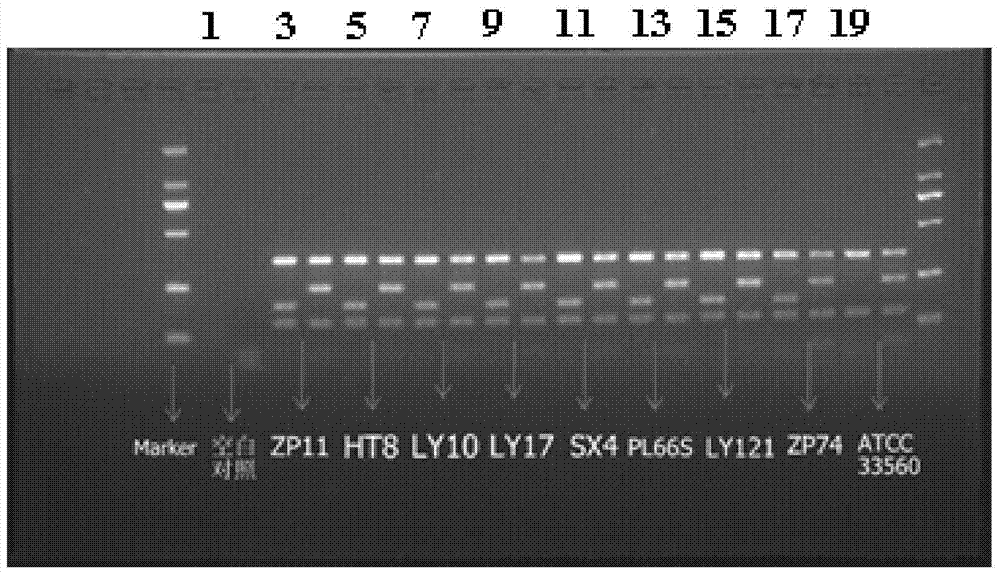
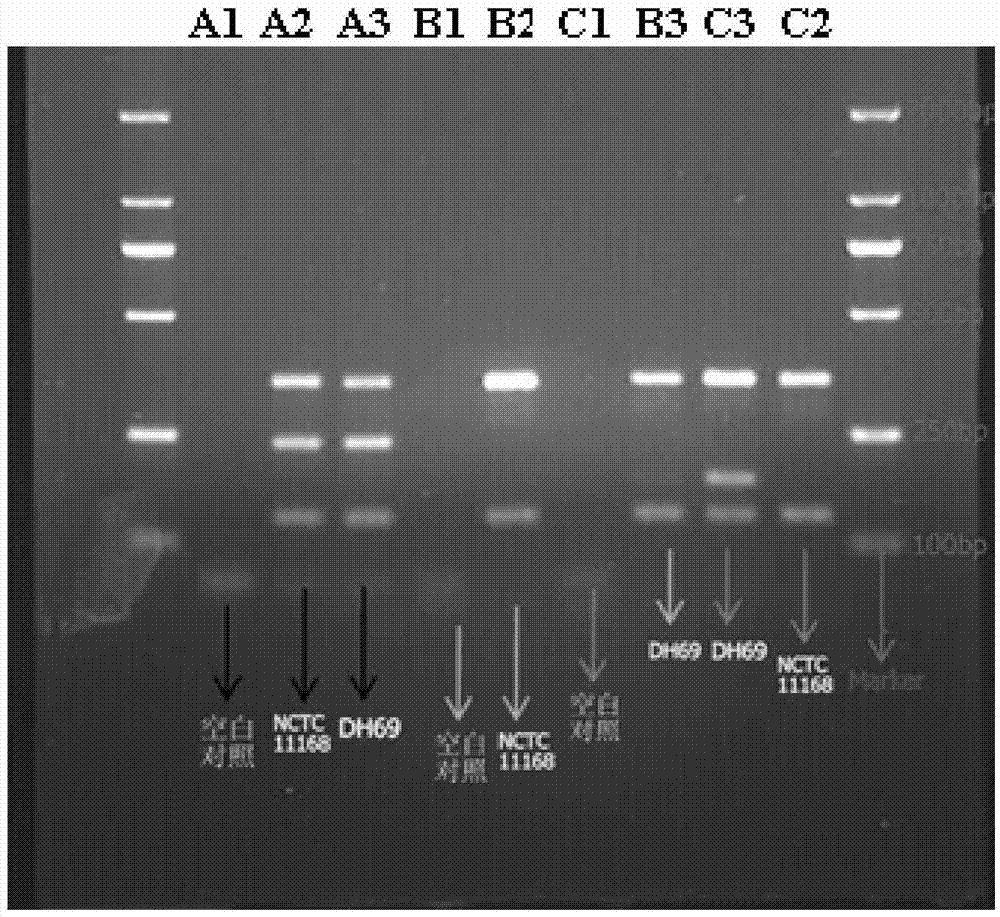
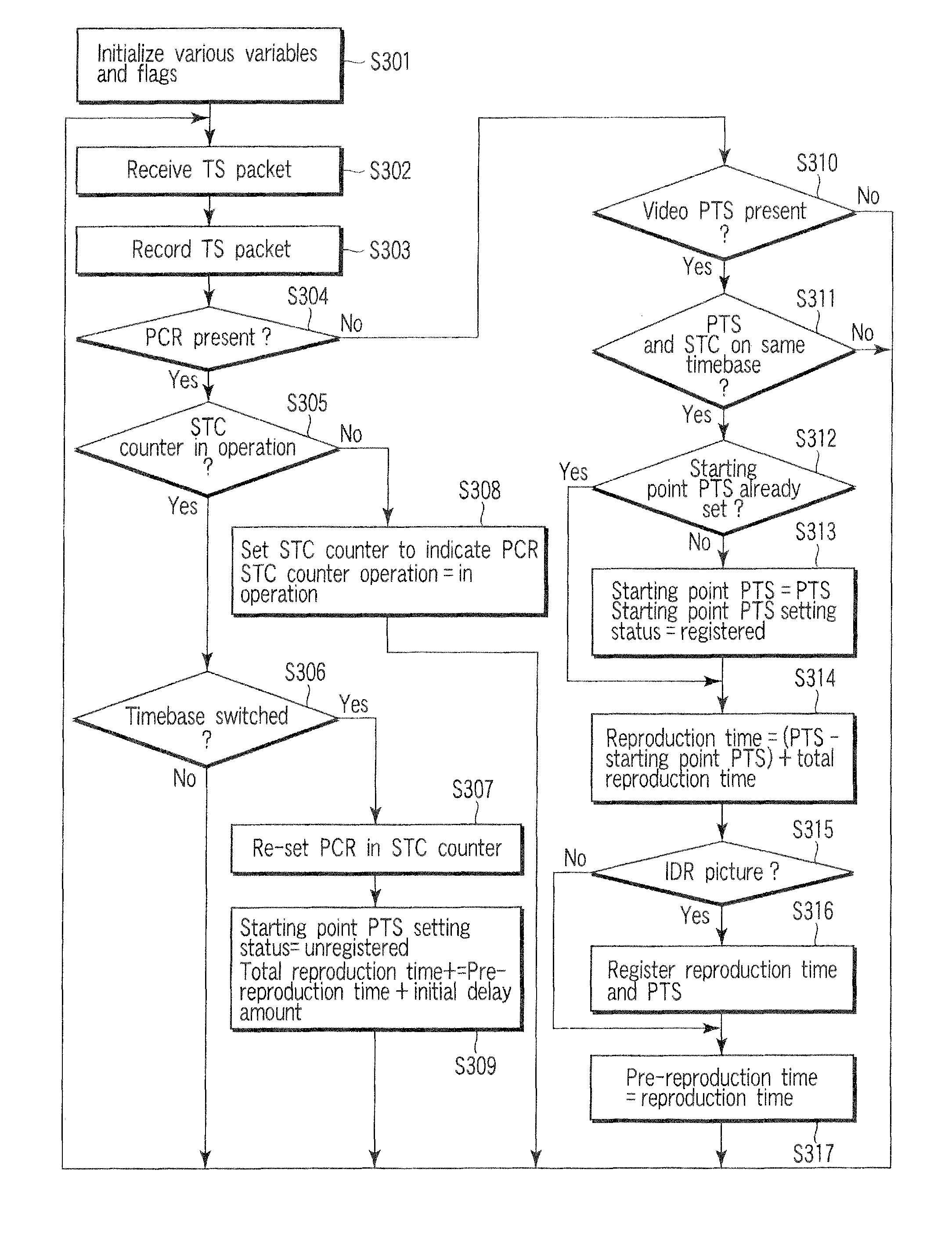
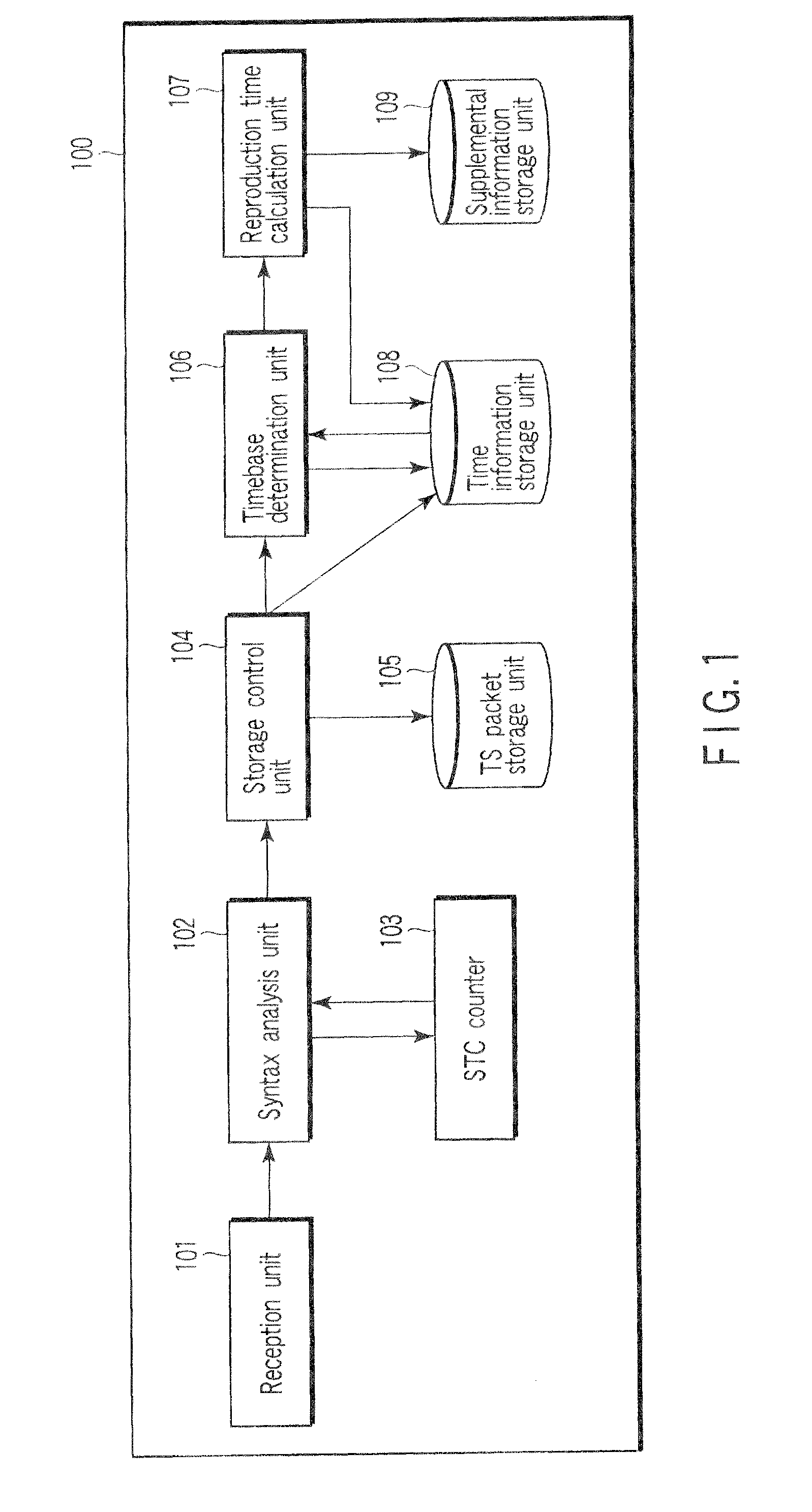
InactiveUS8447159B2Television system detailsAnalogue secracy/subscription systemsReal-time computingAnalysis pcr
A stream recording apparatus includes a reception unit configured to receive a transport stream containing TS packets which include PCRs and PTSs, a first storage unit configured to store the transport stream, an analysis unit configured to analyze the PCRs and the PTSs, a counter configured to count a STC on the basis of one of the PCRs, a calculation unit configured to calculate a reproduction time corresponding to a PTS if the PTS is on a same time base with reference to a STC obtained upon reception of the TS packet having the PTS, and to stop calculating the reproduction time corresponding to the PTSs if the PTSs is on a different timebase, and a second storage unit configured to store the reproduction time in association with the one of the PTSs.
Owner:FUJITSU TOSHIBA MOBILE COMM LTD
Method of determining types I, II, III, IV or V or methicillin-resistant Staphylococcus aureus (MRSA) in a biological sample
ActiveUS8715936B2Simple and sensitive screeningMuch sensitive and accurateAntibacterial agentsBiocidePresent methodStaphylococcus cohnii
Disclosed are diagnostic methods for determining a subtype of methicillin-resistant Staphylococcus aureus (MRSA) in a biological sample of a mammal. Methods include providing a biological sample of the mammal, performing a PCR analysis of the biological sample, and analyzing the PCR amplicons with respect to their sizes so as to determine for type I, type II, type III, type IV or type V MRSA that may be present in the biological sample. Further example embodiments include using at least one mecA primer pair and / or using at least one Staphylococcus aureus nuc primer pair in the PCR analysis. Further disclosed are methods for screening populations for MRSA, and methods of treating a mammal testing positive for Type IV MRSA. Also disclosed are kits for determining a MRSA subtype in a mammal and isolated primers that may be used in the present methods and kits.
Owner:MEDICAL DIAGNOSTIC LAB
Method, kit and application for detecting STR locuses of hepatitis B infected related gene
InactiveCN101597651AAccurate measurementMicrobiological testing/measurementChemiluminescene/bioluminescenceCapillary electrophoresisFluorescence
The invention discloses a method, a kit and application for detecting genotypes of two STR locuses of a hepatitis B virus infected related IL-10 gene. The method is STRs-PCR, which comprises the steps of preparation of a template DNA, primer design of two STR locuses, assembly of a PCR amplification system, PCR circulation reaction, capillary electrophoresis, fluorescence detection and analysis of a PCR product. Compared with the prior art, the invention ensures that the two STR locuses of the IL-10 gene are related with the susceptibility of hepatitis B virus infection, determines IL-10.G and IL-10.R as novel related genes of the hepatitis B virus infection, detects the IL-10.G and the IL-10.R of the IL-10 genes by adopting the STRs-PCR method, thereby determining the susceptibility of individuals to the hepatitis B, and providing new genetic counseling contents for the prevention and control of hepatitis B virus infected groups.
Owner:GUIYANG MEDICAL UNIVERSITY
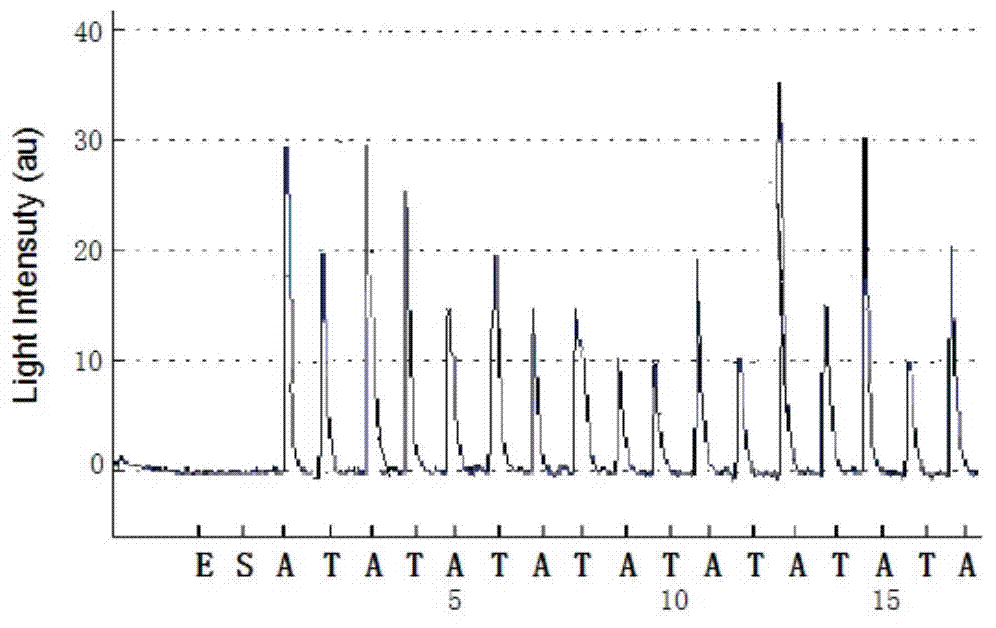
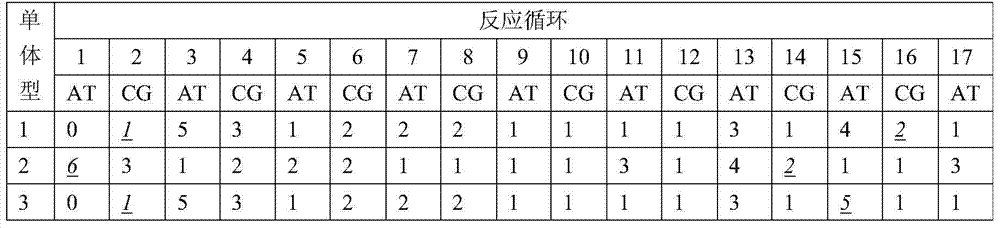
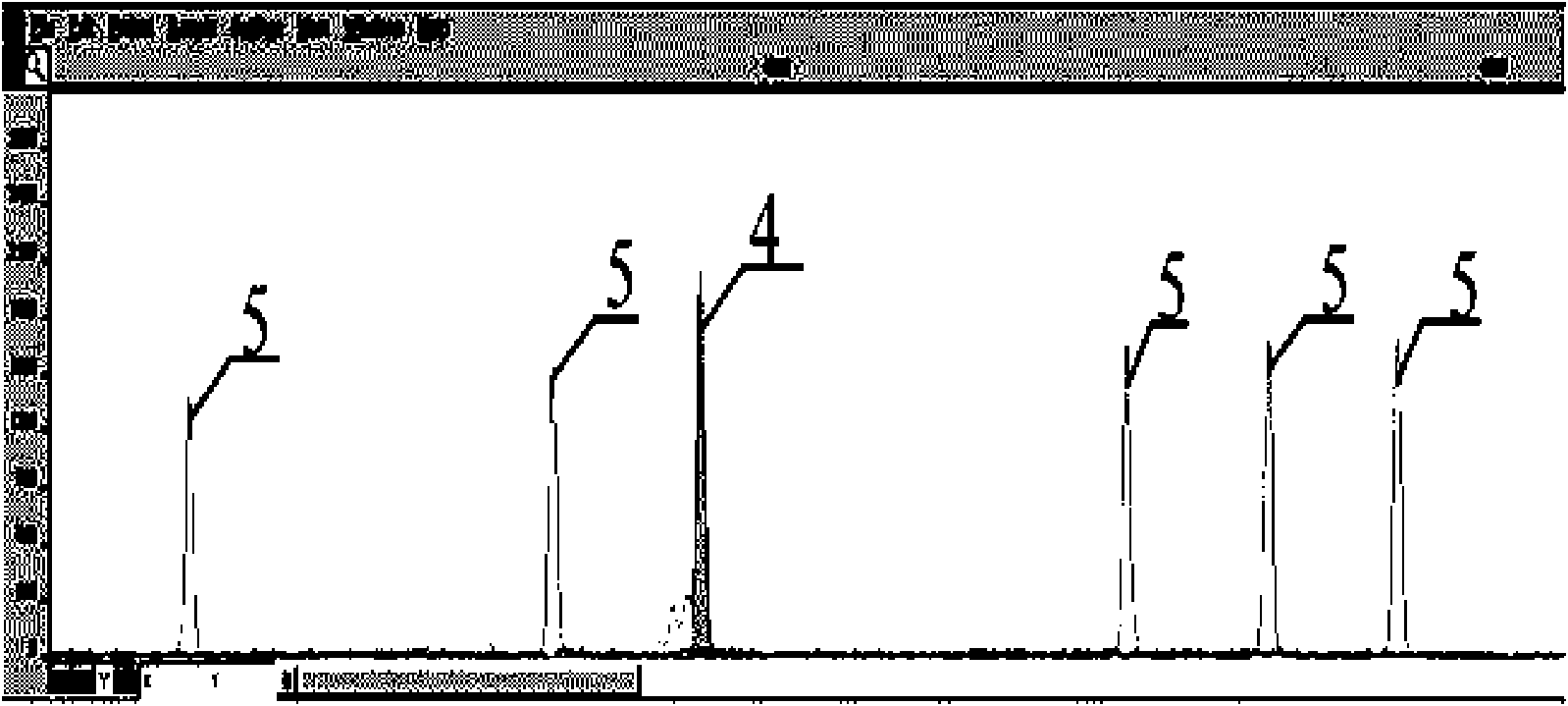
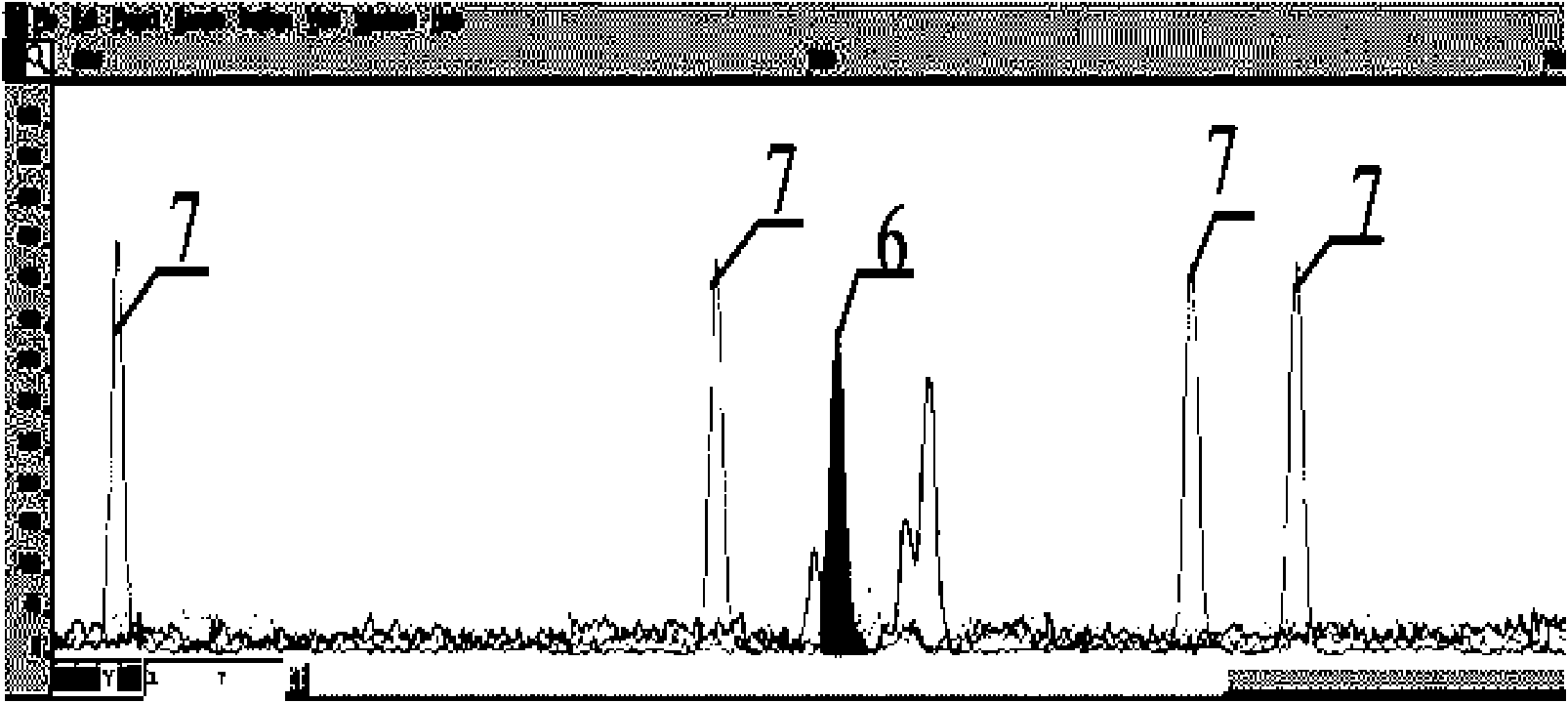
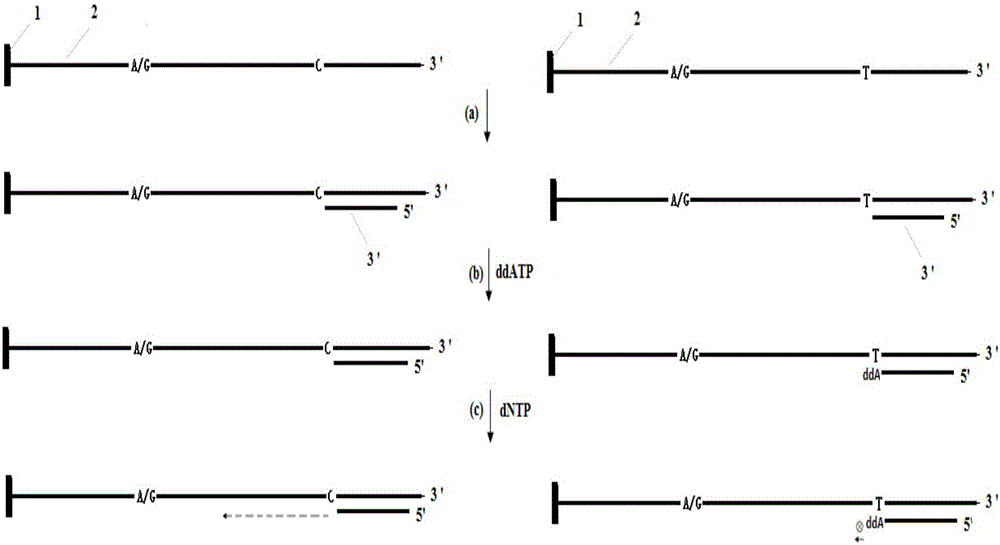
A method for primer selection, sequencing by synthesis, and analysis of haplotypes of PCR products
ActiveCN104313168BHaplotype determinationAnalysis fitMicrobiological testing/measurementAgricultural scienceA-DNA
The invention provides a method for analyzing haplotype of PCR products by primer selection sequencing-by-synthesis. The method is adopted to perform haplotype analysis on two or more PCR products (multiple DNA templates) of site variation (such as SNP and mutation), wherein the PCR products are hybridized by a specific sequencing primer and then are divided into two parts, and a 3' terminus-closed monomer reagent is added to each part; the sequencing primer extends one base with the varied site under the guidance of a DNA template, the 3' terminus of the sequencing primer is closed and cannot extend continuously, and then the sequencing-by-synthesis is performed on the unclosed primers to obtain the sequence information of a specific DNA template, thereby determining the haplotype of PCR products.
Owner:SOUTHEAST UNIV
Polymerase chain reaction detection method of hepatitis B virus genotyping
ActiveCN102140537BSimple and fast operationHigh compliance rateMicrobiological testing/measurementFluorescence/phosphorescenceSerum samplesFluorescence
The invention provides a polymerase chain reaction (PCR) detection method of hepatitis B virus (HBV) genotyping. The method comprises the following steps: a primer is designed according to the entire genome sequence of the HBV genotype; in the existence of the primer, DNAs are extracted from a serum sample to perform PCR amplification in a fluorescence PCR meter, the PCR product is analyzed according to the melting curve, the genotype is judged according to the Tm value of the PCR product and the HBV genotyping can be realized. The PCR melting curve method used in the invention is convenient to operate, the detection result of the method has higher coincidence rate with the detection result obtained by adopting a commercial genotyping kit; and the method can be used in the clinical detection of the HBV genotype. On the basis of the PCR technology, the PCR melting curve method which is to judge the HBV genotype according to the Tm value, is provided in the PCR detection method, thus the HBV genotype can be identified conveniently, rapidly and accurately.
Owner:THE FIRST AFFILIATED HOSPITAL OF FUJIAN MEDICAL UNIV
Reagent system and kit for NDRG4 gene methylation detection and application thereof
InactiveCN106521006AQuantitatively accurateIncreased sensitivityMicrobiological testing/measurementFluorescenceBuffer solution
The invention relates to a reagent system and kit for NDRG4 gene methylation detection and application thereof. The reagent system or the kit comprises specific primers SEQ ID NO.1 and SEQ NO.2 and a specific probe SEQ ID NO.3, and a report group and a quenching group are connected to the probe. The reagent system further comprises a positive standard substance, a negative standard substance, a buffer solution, dNTPs and DNA polymerase. A use method of the kit comprises the steps that 1, methylation treatment is conducted on a sample DNA; 2, a fluorescent quantitative PCR reaction is conducted on the sample DNA obtained after methylation treatment in the step 1 by means of the specific primers SEQ ID NO.1 and SEQ NO.2 and the specific probe SEQ ID NO.3, a fluorescent quantitative PCR reaction is conducted on the positive standard substance and the negative standard substance simultaneously, and the methylation state of a gene is judged by analyzing fluorescence alteration of the PCR process. The preparation method of the kit is simple, the kit is convenient to use and high in speed, and the detection result is accurate, visual and high in sensitivity.
Owner:刘鹏飞
A kit for detecting bovine circovirus and a RT-PCR detection method
InactiveCN109207642AGood repeatabilityRealize real-time monitoringMicrobiological testing/measurementDNA/RNA fragmentationCircovirusRNA extraction
The invention discloses a kit for detecting bovine circovirus, which comprises a primer for detecting bovine circovirus, an RNA extracting reagent, a reverse transcription reagent, a positive controland a negative control, wherein the nucleotide sequence of the primer is an upstream primer P1: 5 '-GGTGCCGTTGTTGTGTCA-3 ', downstream prim P2: 5'-CAGCCCTGAGTTGCCTTATC-3'; Positive control is BCoV, BNoV, BAstV, BRV, BVDV and BKV, negative control is sterile double distilled water. The invention also discloses an RT-A PCR method comprises the following steps: (1) synthesizing the primers for detecting bovine circovirus; (2) extracting RNA from the sample to be tested, and reverse transcribing the extracted RNA as a template to obtain cDNA; (3) PCR amplification reaction is carried out by usingthe cDNA prepared in the step (2) as a template and the primer set synthesized in the step (1); (4) PCR amplification products are analyzed. Simple, practical, economical and efficient, it can greatlyimprove the detection efficiency and save the detection time.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY MEDICINE HENAN ACAD OF AGRI SCI
Method for quickly detecting specific PCR (polymerase chain reaction) products on basis of analysis on PCR byproduct pyrophosphoric acid and application of method
PendingCN109797195ALow background valueHigh detection sensitivityMicrobiological testing/measurementBiologyLarge sample
The invention discloses a method for quickly detecting specific PCR (polymerase chain reaction) products on the basis of analysis on a PCR byproduct pyrophosphoric acid and application of the method.The method includes steps of extracting sample genomes for PCR; (2), designing at least one PCR primer and substituting alpha-sulfo-deoxyadenosine triphosphate for deoxyadenosine triphosphate and carrying out PCR amplification; (3), analyzing the pyrophosphoric acid in PCR amplification products; (4), judging whether the PCR is carried out or not according to analysis results and detecting the content of to-be-detected DNA (deoxyribonucleic acid) in samples. The method and the application have the advantages that whether the samples contain sample DNA or not and the content of the sample DNA can be accurately detected by the aid of the method, and the detection limit of the method can reach a plurality of copies; the method for detecting and analyzing the specific PCR products is high in sensitivity, the specific PCR products can be quickly detected by the aid of the method, operation processes are simple and are easy to implement, and large sample specific PCR products can be quicklydetected by the aid of the method; the method can be used for quickly detecting microorganisms in the industry of food, sanitation and the like and also can be used for analyzing other nucleotide sequences with specific PCR.
Owner:SOUTHEAST UNIV
A method for analyzing the haplotype of PCR products by asynchronous synthesis and sequencing of two nucleotides
ActiveCN104762406BBroaden the scope of analysisEasy to operateMicrobiological testing/measurementSynthesis methodsSynthetic DNA
The invention discloses a method for analyzing PCR product haplotype by synchronous synthesis and sequencing of two nucleotides. Through real-time synthesis and sequencing of two nucleotides, two different nucleotides are added to each sequencing reaction at the same time, and the synthesis and sequencing of different DNA templates in the PCR product are not synchronized, so that the number of different bases on two adjacent SNP sites Information is provided by the different sequencing reactions. Based on the sequencing information at different SNP sites obtained from these asynchronous synthesis sequencing reactions, association analysis was performed to determine the haplotype of the PCR product. The invention broadens the analysis range, is simple to operate, and can reduce the analysis cost; it is suitable for the haplotype analysis of multiple mixed samples, can directly measure the ratio of each haplotype in the mixed sample, and can be used to find, Screening for nucleic acid markers.
Owner:SOUTHEAST UNIV
Method, kit and application for detecting STR locuses of hepatitis B infected related gene
InactiveCN101597651BMicrobiological testing/measurementChemiluminescene/bioluminescenceCapillary electrophoresisFluorescence
The invention discloses a method, a kit and application for detecting genotypes of two STR locuses of a hepatitis B virus infected related IL-10 gene. The method is STRs-PCR, which comprises the steps of preparation of a template DNA, primer design of two STR locuses, assembly of a PCR amplification system, PCR circulation reaction, capillary electrophoresis, fluorescence detection and analysis of a PCR product. Compared with the prior art, the invention ensures that the two STR locuses of the IL-10 gene are related with the susceptibility of hepatitis B virus infection, determines IL-10.G and IL-10.R as novel related genes of the hepatitis B virus infection, detects the IL-10.G and the IL-10.R of the IL-10 genes by adopting the STRs-PCR method, thereby determining the susceptibility of individuals to the hepatitis B, and providing new genetic counseling contents for the prevention and control of hepatitis B virus infected groups.
Owner:GUIYANG MEDICAL UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com