Method for quickly detecting specific PCR (polymerase chain reaction) products on basis of analysis on PCR byproduct pyrophosphoric acid and application of method
A detection method and pyrophosphate technology, applied in the biological field, can solve the problems of low copy number of DNA template and cannot be effectively detected, and achieve the effects of accurate qualitative and quantitative analysis, easy implementation and rapid detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
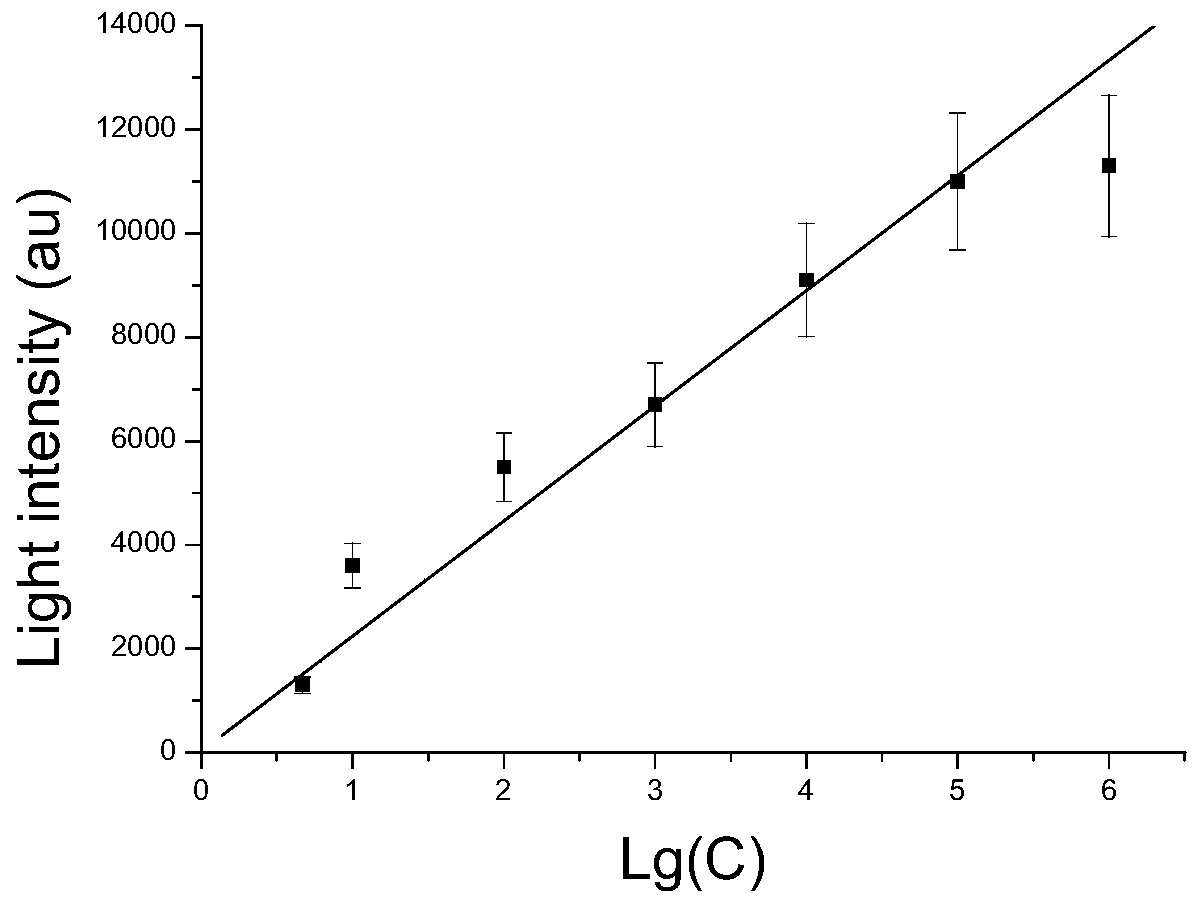
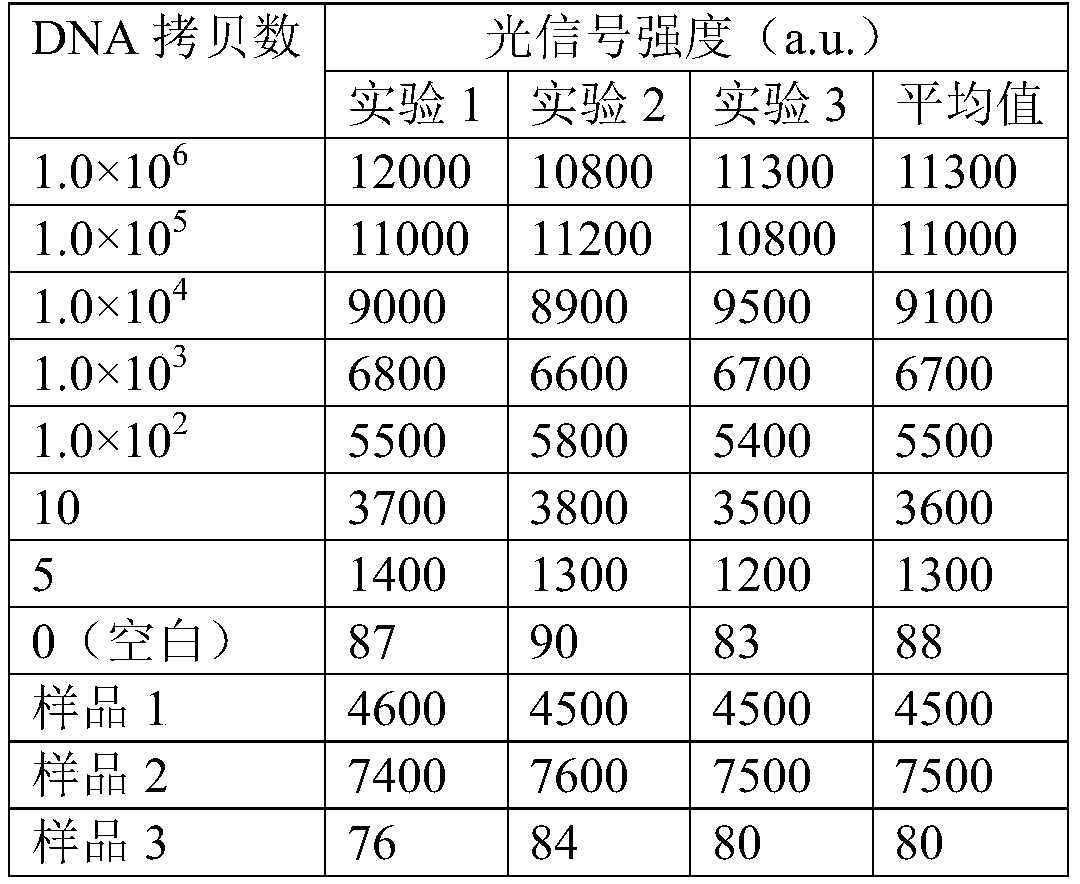
[0035] Detection of Salmonella in Chicken
[0036] 1. Genomic DNA sample preparation: Use PBS (pH=7.2) to wash the bacteria on the surface of the chicken, and then use the bacterial DNA extraction kit (QIAamp fast DNA stool mini kit, Qiagen) to extract the washed bacterial DNA, and store at low temperature spare.
[0037] 2. PCR primer design and PCR reaction
[0038] For the characteristic nucleic acid sequence fragment of Salmonella, the Salmonella invA gene is a gene encoding a secreted virulence protein, which is closely related to the pathogenicity of the bacterium and is unique to the genus Salmonella. The forward primer and reverse primer were designed according to GenBank accession no.M76445.1 :
[0039] SEQ ID NO.1 is forward primer F: 5'-CATCTGTCTCGCCTCCTG-3',
[0040] SEQ ID NO.2 is the reverse primer R: 5'-GGGCCATTTGTCTACTCATA-3';
[0041] (Search the sequence of the amplified gene in the GeneBank database, and submit it to the primer design software Primer5.0 ...
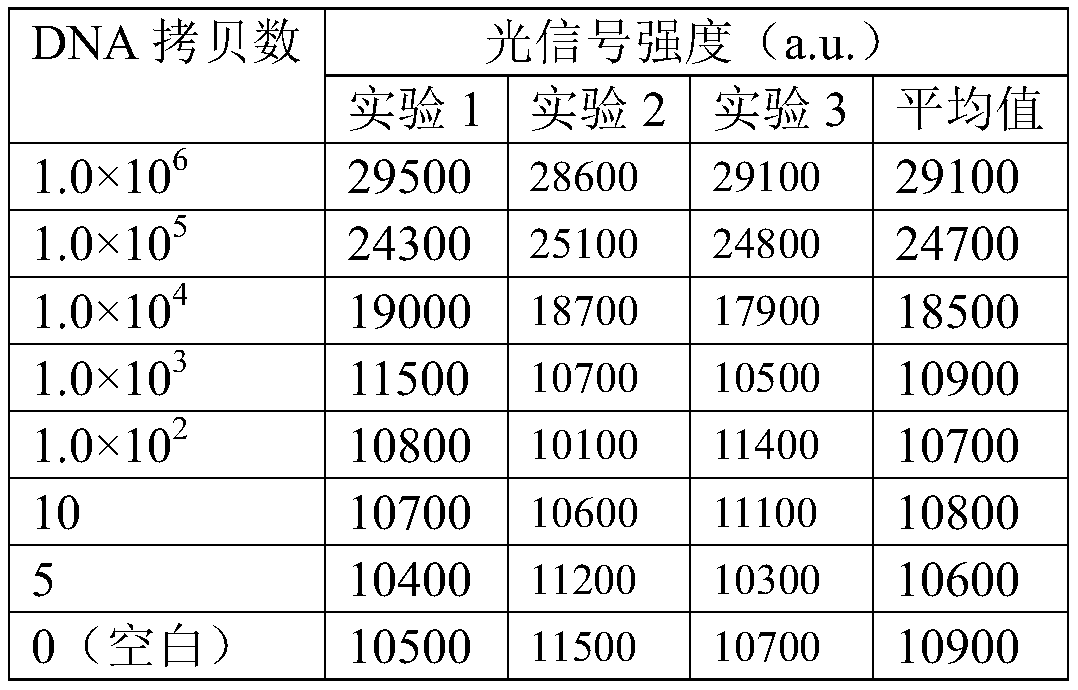
Embodiment 2
[0101] Example 2 adopts the same method as Example 1, except that the pyrophosphate chemiluminescence analysis method uses a pyrophosphate detection kit purchased by SIGMA-ALDRICH Company, and the detection result is similar to that of Example 1.
Embodiment 3
[0103] Detection method of rs11053646 SNP site based on analysis of rolling circle amplification by-product pyrophosphate
[0104] 1. DNA sample extraction and processing: Peripheral blood samples were extracted with traditional protein kinase K and phenol / chloroform extraction methods to extract genomic DNA from peripheral blood. Genomic DNA was digested with restriction enzyme Ase I (New England Biolabs, United States).
[0105] 2. Ring connection reaction:
[0106] (1) Ligation reaction of loop 1: 10 μL reaction volume contains 100 ng of restriction endonuclease Ase I digested genomic DNA, 1 nM of artificially synthesized open loop DNA sequence SEQ ID NO.3: (5'-PO 4 3— AGCCAAGAGAAGTGCGTAACGGTGTGATATGAGCGTGATCGTGCCTTGTCATTCGGGAAACTGGGAAAA G- 3'), 2U Ligase (Epicentre Technologies), 26mmol / L tris-HCl (pH=8.3), 32.5mmol / L KCl, 13mmol / l MgCl 2 , 0.65mmol / L NAD, and 0.013% X-100. Then the reaction was carried out at 95°C for 5 minutes, 40 thermal cycles: denaturation at 9...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com