Primer for specifically detecting human genome DNA and application thereof
A genome and human-derived technology, applied in the direction of recombinant DNA technology, DNA/RNA fragments, microbial measurement/testing, etc., can solve the problems of insufficient sequence sensitivity, inconsistent results, inaccurate results, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2
[0079] The sensitivity of embodiment 2SRGAP2 qPCR detection method
[0080] The primers and probes for the specific fragments of human SRGAP2 and FOX2A were designed and synthesized, and the primers and probes for SRGAP2 and FOX2A were as described in Example 1.
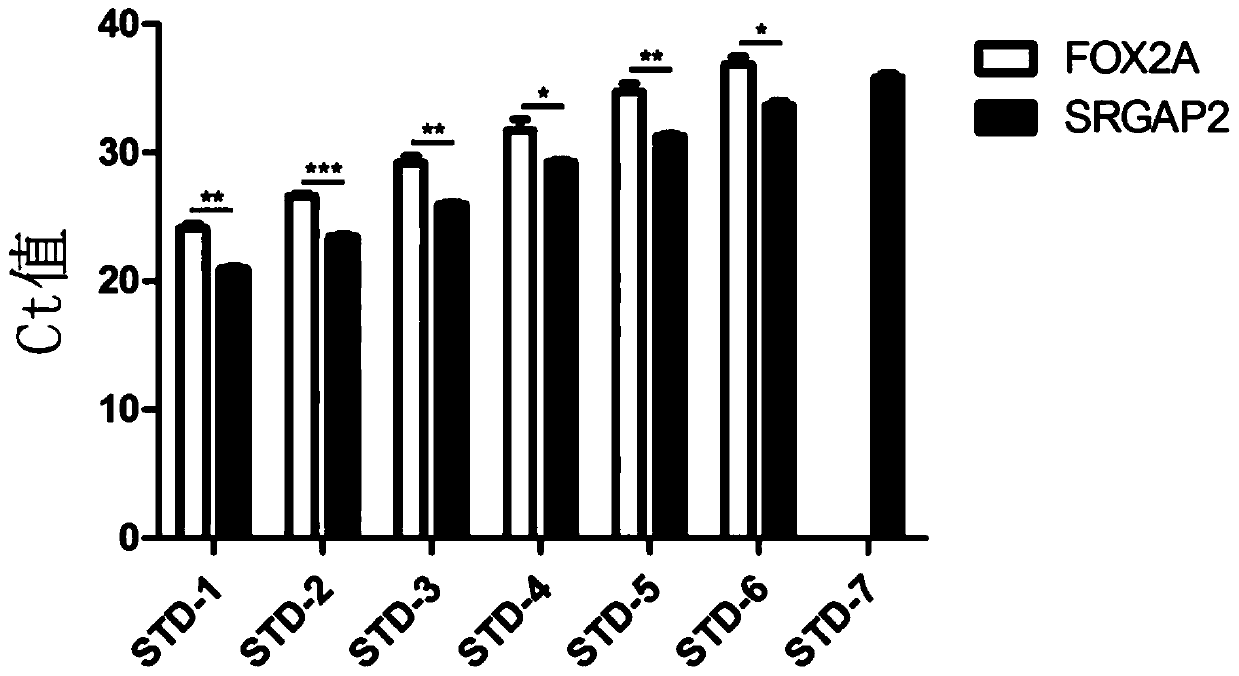
[0081] Genomic DNA extracted from human mesenchymal stem cells (MSC) was used as a standard, starting with 100ng of human mesenchymal stem cell genomic DNA, using New Zealand rabbit liver tissue genomic DNA as a diluent, and 5-fold gradient dilution to prepare 7 concentration samples (STD -1 to STD-7, wherein the content of human-derived genomic DNA is 100000pg, 20000pg, 4000pg, 800pg, 160pg, 32pg, 6.4pg per 5μl sample), and the primers and probes of SRGAP2 and FOX2A were used to pair these 7 Concentration samples were subjected to qPCR experiments to compare the specificity and sensitivity of the two primers. The result is as figure 2 Shown, where *p<0.05; **p<0.01. The results showed that the specificity of t...
Embodiment 3
[0082] Example 3 Formulate the standard curve of SRGAP2 qPCR detection method
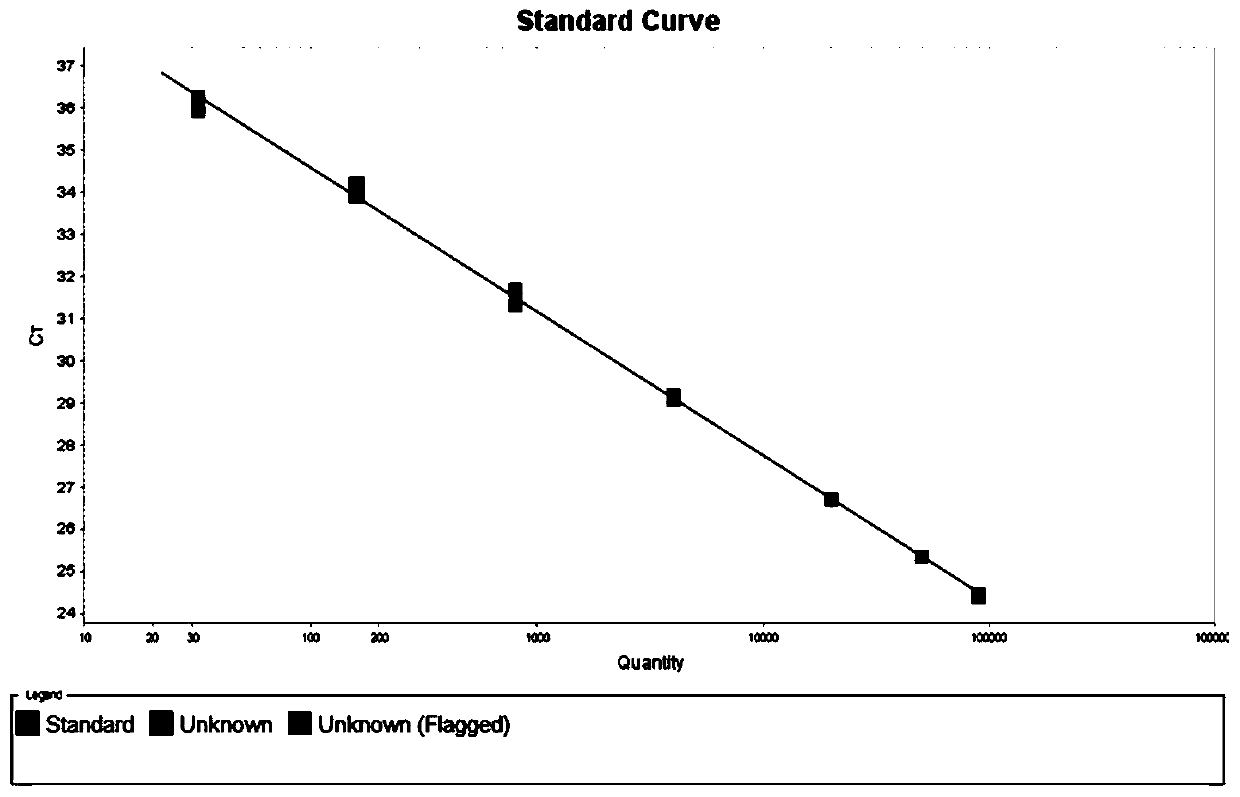
[0083] Using New Zealand rabbit liver genomic DNA as the diluent, the concentration of human mesenchymal stem cell genomic DNA was serially diluted, and qPCR detection was performed with the SRGAP2 primer probe as described in Example 1 until the lowest limit of stable detection was 32.00pg / 5μl is set as the lower limit of detection (or lower limit of quantification); the highest limit that can be stably detected is 90000.00pg / 5μl, which is set as the upper limit of detection (or upper limit of quantitation); concentrations higher than the upper limit of detection can also be detected , but false positive results may occur.
[0084] With the upper detection limit and lower detection limit as the limit, human mesenchymal stem cell genomic DNA and New Zealand rabbit liver genomic DNA were mixed to prepare samples of standard concentration, and qPCR detection was performed using the SRGAP2 primer p...
Embodiment 4
[0094] Example 4 SRGAP2 qPCR method precision, accuracy detection
[0095] The lower limit of quantification (LLOQ, Lower limit of quantification), low concentration quality control (LQC, Low Quality control), high concentration quality control (HQC, High Quality control), Samples with 5 concentrations including middle concentration quality control (MQC, Middle Quality control) and upper limit of quantification (ULOQ, Upper limit of quantification) were subjected to qPCR with the SRGAP2 primer probe as described in Example 1. The concentration of each sample was as described for the corresponding sample in Example 3. Amplification curve such as Figure 4 shown.
[0096] The intra-assay and inter-assay precision and accuracy of samples with different concentrations were investigated. Accept the Guidelines for the Validation of Quantitative Analytical Methods for Biological Samples in the Standard Reference Pharmacopoeia (14-16), as follows:
[0097]
[0098] Remark:
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com