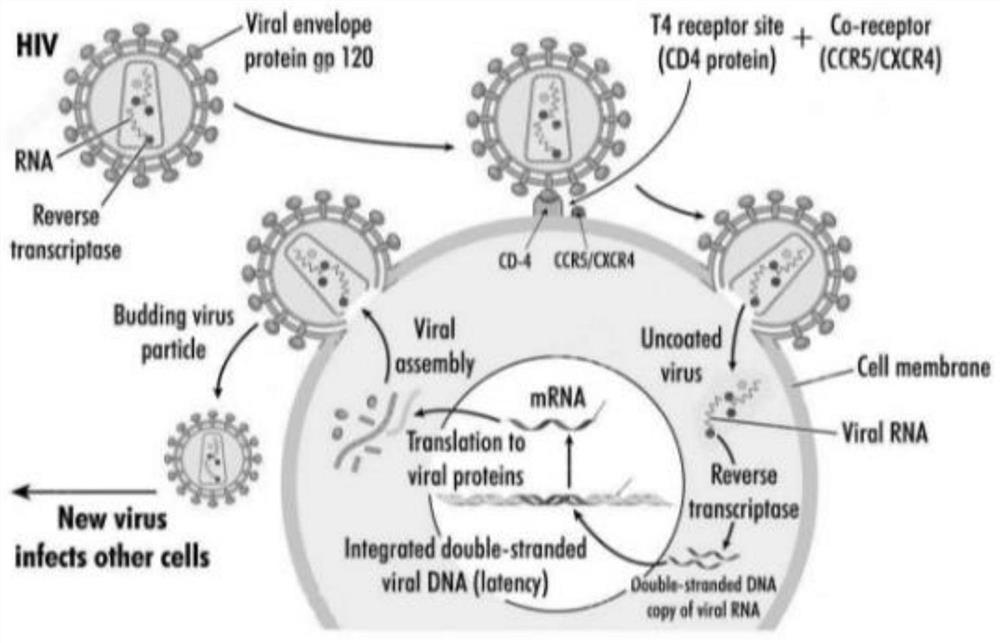
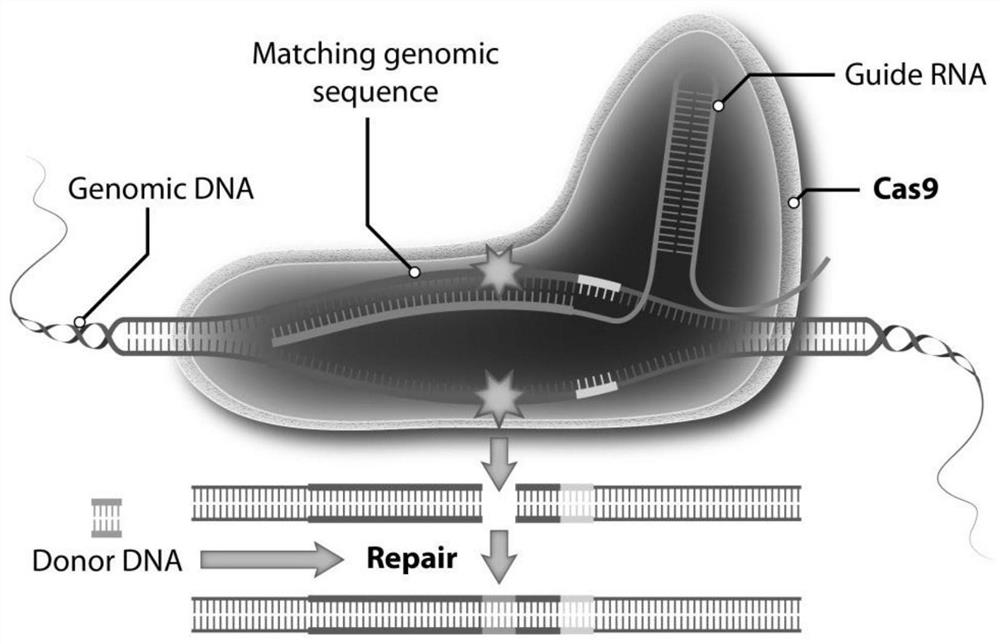
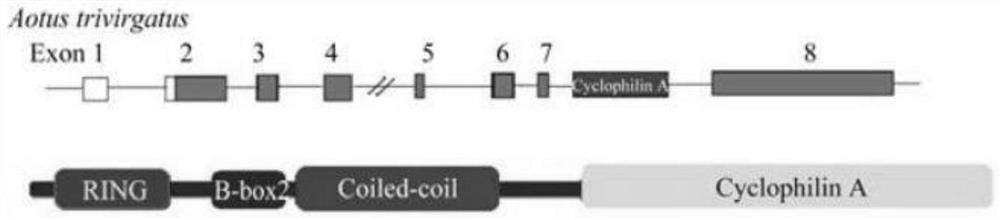
A method for site-specific editing of ccr5 gene
A gene editing technology, applied in genetic engineering, plant genetic improvement, botanical equipment and methods, etc., can solve problems such as uncontrollable, risky, fatal safety, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1 High-efficiency gRNA screening of CRISPR-Cas9 knockout CCR5 gene
[0039] 1. gRNA preparation
[0040] (1) Design a 20nt gRNA sequence according to the sequence of the CCR5 gene (including the exon and the intron sequence adjacent to the exon), the 3'-end of the target sequence of the gRNA on the CCR5 gene is NGG;
[0041] (2) Synthesize the sense strand and antisense strand of the target sequence with cohesive ends respectively (cacc is added to the 5'-end of the sense strand, if the first nucleotide at the 5'-end of the sense strand is not guanine G, add cacc to the sense strand Add caccG at the 5'-end of the antisense strand; add aaac at the 5'-end of the antisense strand, if the first nucleotide at the 5'-end of the sense strand is not guanine G, add C at the 3'-end of the antisense strand );
[0042] (3) The above-mentioned sense strand and antisense strand were mixed, treated at 90°C, cooled naturally to room temperature for annealing treatment, and do...
Embodiment 2
[0068] Example 2 Construction of a donor vector for site-specific integration of the hTRIM5 / CypA gene at the CCR5 site
[0069] (1) Acquisition of the upstream homology arm F86-F of CCR5F86
[0070] Construction of pDonor-TALEN-EGFP-hCCR5F86-F expression vector
[0071] 1. PCR amplification of the target fragment F86-F;
[0072] Primers F86-SnaB I-L / F86-Sal I-R were used to amplify the F86-F target fragment using the HepG2 cell genome as a template.
[0073] F86-SnaB I-L (SEQ ID NO: 3):
[0074] 5′-gttctagtggttggctacgtagcaccatgcttgacccagtttc-3′
[0075] F86-Sal I-R (SEQ ID NO:4):
[0076] 5′-tagcttatcgataccgtcgacGTCACCACCCCAAAGGTGAC-3′
[0077] The resulting PCR product sequence contains the DNA sequence SEQ ID NO:5 of the F86 upstream homology arm F86-F:
[0078]gcaccatgcttgacccagtttcttaaaattgttgtcaaagcttcattcactccatggtgctatagagcacaagattttatttggtgagatggtgctttcatgaattcccccaacagagccaagctctccatctagtggacagggaagctagcagcaaaccttcccttcactacaaaacttcattgcttggccaaaaagagagttaattCAA...
Embodiment 3HI
[0134] The transformation of embodiment 3 HIV-1 susceptible cell lines
[0135] 1. Culture Hela-CD4 (X4 tropism-susceptible cell line) and TZM-bl (R5 tropism-susceptible cell line) cells on the wall of 24-well plate, the plating density is 1.2×10^5cells / well, use DMEM medium (Add 10% FBS) culture;
[0136] 2. Extract the plasmid pDonor-EF1α-hTRIM5 / CypA-EGFP-hCCR5F86 ( Figure 10 ) and pX458-F86;
[0137] 3. Using Lipofectamine 3000Transfection Reagent, the above two plasmids were co-transformed into Hela-CD4 and TZM-bl cell lines;
[0138] 4. From the fifth day, add G418 with a final concentration of 800μg / ml for selection, passaging once every 2-3 days, and replace each time with fresh DMEM medium containing 800μg / mL G418;
[0139] 5. After 12 days of drug screening, cells were digested, sorted by flow cytometry and inoculated into 96-well plates;
[0140] 6. Expand the culture of cells sorted into 96-well plates, select cell lines that stably and uniformly express green ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com