High-resolution-ratio dissolution curve detection method for mutation of OsNramp5 gene specific site
A melting curve and high-resolution technology, which is applied in the field of high-resolution melting curve detection, can solve the problems of time-consuming and labor-intensive detection, and achieve the effects of fast detection time, cost saving and efficiency improvement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] In the following examples, the experimental methods that do not indicate specific conditions are generally in accordance with conventional conditions, such as the book "Molecular Cloning Experiment Guide (3rd Edition)" (J. Sambrook, written by D.W. Russell; Huang Peitang et al. translation; Science Press Society, 2008), or in accordance with the conditions recommended by manufacturers of reagents or equipment manufacturers.
[0039] Embodiment 1, the creation of a specific site mutation of OsNramp5 gene
[0040] The specific site of the OsNramp5 gene was mutagenized by the CRISPR / Cas9 system, and the steps were as follows:
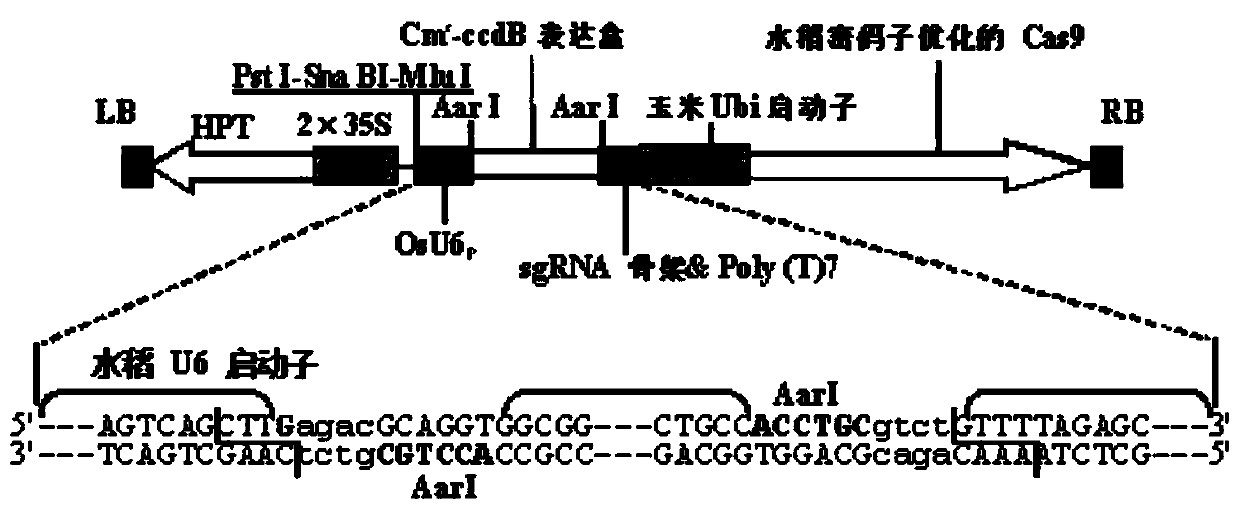
[0041] 1. CRISPR vector construction
[0042] CRISPR / Cas9 vector construction. The CRISPR / Cas9 vector used in this study was self-constructed, and the vector was the binary vector pCUbi1390 (obtained by adding the corn Ubiquitin3 promoter to the pCAMBIA1390 backbone) [Peng H, ZhangQ, Li Y, et al. Aputative leucine-rich repeat receptor kinase , Os...
Embodiment 2
[0057] Example 2. Using the high-resolution melting curve detection method to detect mutations at specific sites in the OsNramp5 gene
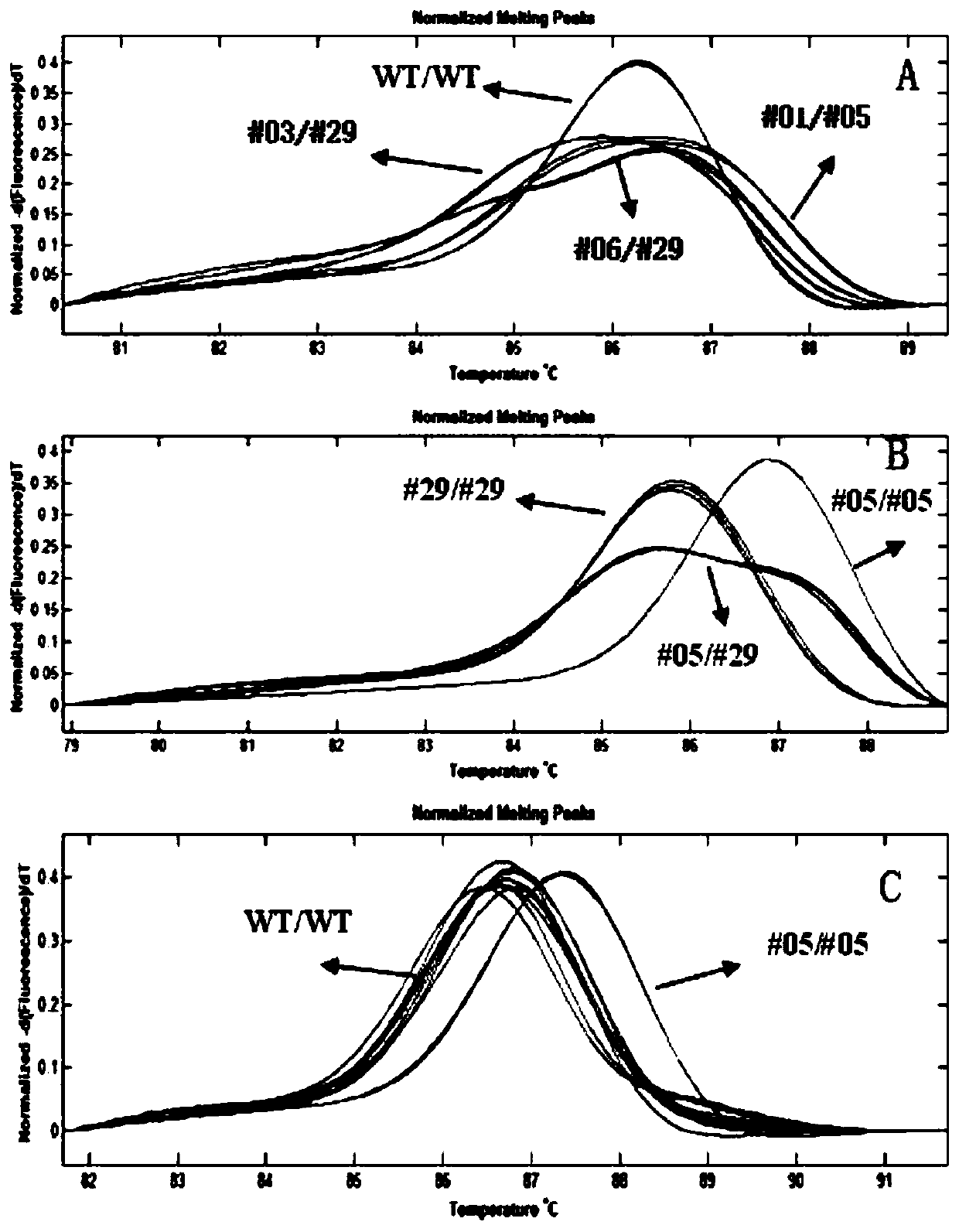
[0058] Using the high-resolution melting curve method provided by the present invention, the transformed seedling T0 generation plants obtained by the method of Example 1 were analyzed, and compared with the sequencing results, it was found that the method provided by the present invention can identify almost all heterozygous or diisozygous For the genotype of the allele mutation, see typical test results image 3 (A).
[0059] For the mutant T1 generation plants obtained in Example 1, after using the leaf segment hygromycin soaking method to screen out individual plants that are not resistant to hygromycin, that is, do not contain transgenic components such as resistance markers, use the high-resolution method described in the present invention Homozygous identification of mutant individual plants was carried out by the rate melting curve de...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com