CHO cell line with GS gene knockout and preparation method and application thereof
A cell line and gene technology, which is applied in the field of CHO cell line with GS gene knockout and its preparation, can solve the problems of cell resource consumption, cell line canceration, cell genome cutting damage, etc., and achieve faster screening efficiency, simple and efficient operation Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Example 1: Gene editing vector construction
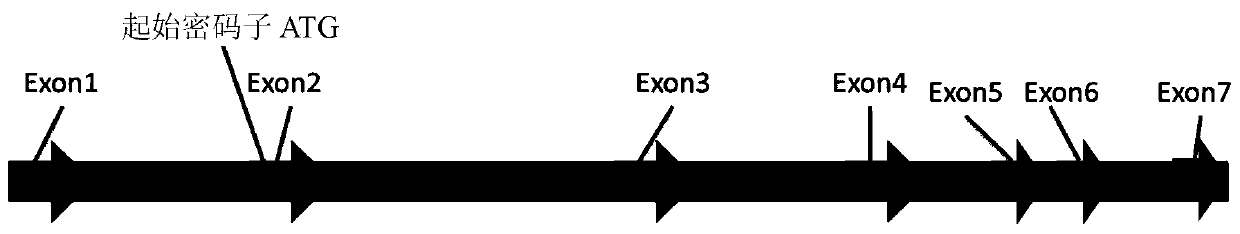
[0071] The GS gene of CHO cells consists of 7 exons, of which the coding sequence (CDS) encoding and expressing the GS protein is located in exons 2-7 (see figure 1 ), and its normal function plays a vital role in cell growth and survival. The partial sequence of exons 2 to 7 of the GS gene is shown below (the CDS sequence is marked with an underline "_____"):
[0072] SEQ ID NO.1: Exon 2
[0073] CCCCTTCAGAGTAGATGTTAATGAAATGACTTTTGTCTCTCCAGAGCACCTTCCACC ATGGCCACCTC AGCAAGTTCCCACTTGAACAAAAACATCAAGCAAATGTACTTGTGCCTGCCCCAGGGTGAGAAAGTCCAAGCCATG TATATCTGGGTTGATGGTACTGGAGAAGGACTGCGCTGCAAAACCCGCACCCTGGACTGTGAGCCCAAGTGTGTAG AAG GTGAGCATGGGCAGGAGCAGGACATGTGCCTGGAAGTGGGCAAGCAGCCTGAGATTTGACCTTCCTTCTGTTTTG;
[0074] SEQ ID NO.2: Exon 3
[0075] GATATACATGCAAGTAAAACACCCCTACACACATAAAAATAAATACGTCTTTCTTAAAAGTTAATTTCCATCTTTATTTGGCCCAG AGTTACCTGAGTGGAATTTTGATGGCTCTAGTACCTTTCAGTCTGAGGGCTCCAACAG TGACATGTATCTCAGCCCTGTTGCCATGTTTCG...
Embodiment 2
[0096] Example 2: GS gene editing of CHO-S cells and isolation and identification of monoclonal cells
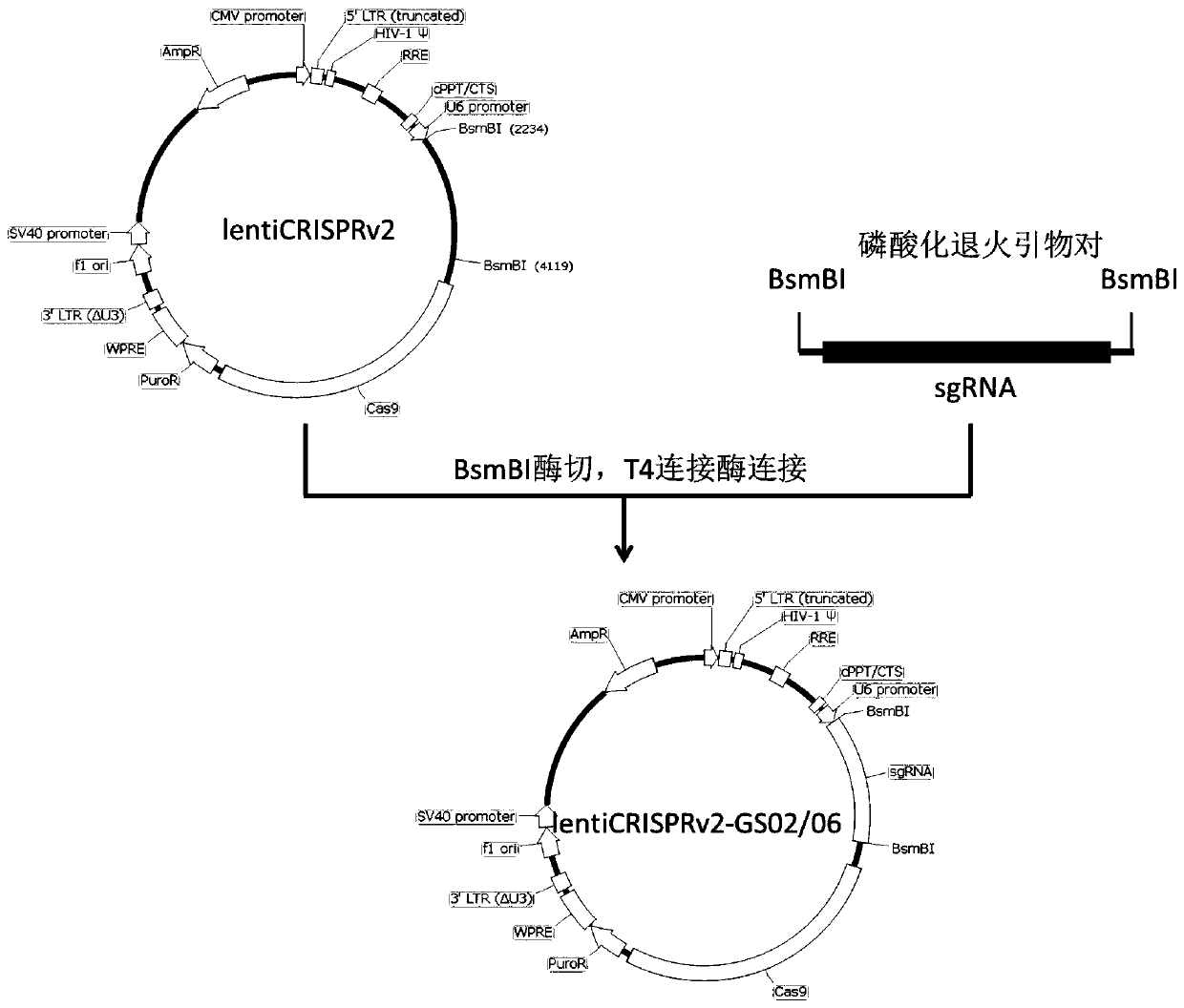
[0097] (1) Plasmid extraction: Use the plasmid purification kit (PureLink TM HiPure Plasmid Filter Midiprep Kit, Thermo Fisher Scientifi), referring to the method in the manual, extracted lentiCRISPRv2-GS02 and lentiCRISPRv2-GS06 plasmids to obtain high-purity, endotoxin-free high-concentration plasmids.
[0098] (2) Cell culture: resuscitated CHO-S (cGMP Banked, ) suspension cells were subcultured in CD FortiCHO complete medium (containing 8mM L-glutamine) in a conventional subculture method, and LentiCRISPRv2-GS02 and LentiCRISPRv2-GS06 transfection experiments were carried out after subculture for 3 generations.
[0099] (3) Cell transfection:
[0100] (A) 22-24 hours before transfection, the CHO-S cells were mixed with (5~6)×10 5 Cells / mL, subculture in 30ml CD FortiCHO complete medium; on the day of transfection, the cell density should be (1.2~1.5)×10 6 cells / ml, ...
Embodiment 3
[0142] Example 3: Determination of the base sequence of the GS gene edited by CRISPR / Cas9
[0143] (1) No. 1, No. 2, and No. 3 (3 GS- / - monoclonal cell lines without sgRNA integration gene, Cas9 integration gene, and puromycin resistance) extracted in Example 2) monoclonal cell lines The genome is used as a template, and the exon 2 and exon 6 CDS sequences of the GS gene are amplified by PCR. The primer sequences and PCR reaction systems are as follows:
[0144] GS02 forward primer (GS02F): 5'-CCCCTTCAGAGTAGATGTTAATGAA-3' (SEQ ID NO.18);
[0145] GS02 reverse primer (GS02R): 5'-CAAAACAGAAGGAAGGTCAAATCTC-3' (SEQ ID NO.19);
[0146] GS06 forward primer (GS06F): 5'-TATGGACTCTGATTCTTCACTG-3' (SEQ ID NO.20);
[0147] GS06 reverse primer (GS06R): 5'-ATGAGAATAAAGATGGCTCCAG-3' (SEQ ID NO. 21).
[0148] 50 μl reaction system: 3 μl (100ng) of template genomic DNA, 2 μl of primer GS02F at a concentration of 5 μM, 2 μl of primer GS02R at a concentration of 5 μM, 25 μl of PrimerSTAR Max...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com