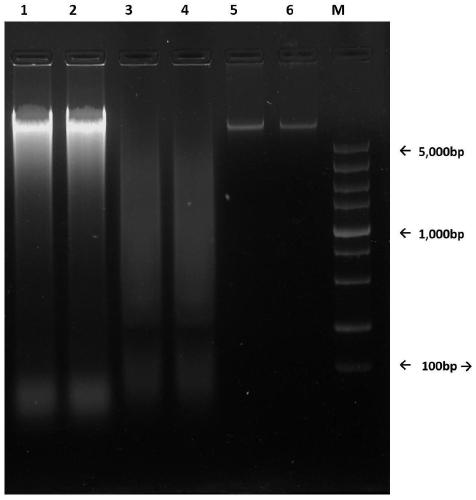
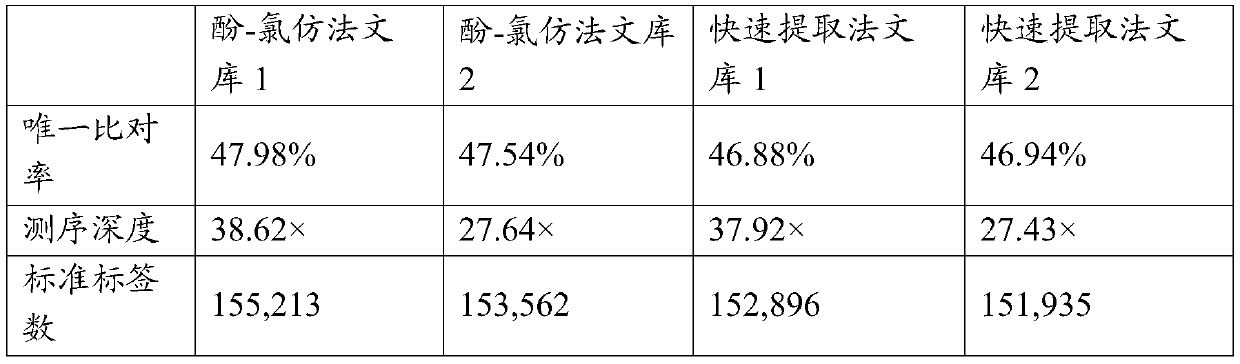
Trace shellfish DNA extraction method for high-throughput sequencing library preparation
A technology for sequencing libraries and extraction methods, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of inability to guarantee the quality of high-throughput sequencing library preparation, affecting endonuclease activity, deformity damage, etc. Ensuring high-quality library preparation, reduced DNA extraction time, and easy and fast operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] A method for extracting trace shellfish DNA for high-throughput sequencing library preparation, specifically comprising the following steps:
[0033] (1) Add 99.5 μl of cell lysate (100 mM NaCl, 10 mM Tris-HCl and 1 mM EDTA, pH=8.0) into a 1.5 ml EP tube, and then add 0.5 μl of proteinase K at a concentration of 20 mg / ml to the lysate , adjusted so that the final proteinase K enzyme concentration of the final mixture reached 0.1 μg / μl to prepare a mixture.
[0034] (2) Put 5.0 mg of gill filaments of isolated Ezo scallop into the EP tube.
[0035] (3) Put the above-mentioned EP tube into a 37-65°C water bath for 1.0h for digestion. This example is a 56°C water bath. In order to promote cell lysis and protein digestion, take out the EP tube and mix it upside down every 30 minutes.
[0036] (4) After digestion, absorb the supernatant and dilute it with triple steamed sterilized water at a ratio of 1:49. The diluent is the scallop genome DNA extracted by the present inve...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com