Humanized anti-PSMA single-chain antibody and application thereof
A single-chain antibody, humanized technology, applied in the field of biomedicine, can solve problems such as immune damage, and achieve the effect of improving effectiveness, small molecular weight, and low immunogenicity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
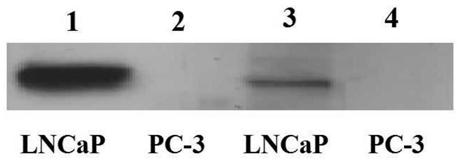
Embodiment 1
[0015] Example 1: Enrichment and screening of phage library using PSMA antigen
[0016] 0.05mol / L carbonate buffer diluted PSMA recombinant protein to 10μg / mL antigen coating solution. Coat the immunotube with 1mL of antigen coating solution and store overnight at 4°C. The next day, the coating solution was discarded, 2.5% skimmed milk powder TBST solution was added, the immunotube was filled, and the tube was sealed at 37°C for 2 hours. After blocking, discard the blocking solution and rinse with PBST 10 times. Add 1 mL of the phage single-chain antibody library whose titer has been determined to the immunotube (the library capacity is about 10 11 CFU), incubated at 37°C for 2h. After the incubation of the antibody library, the phage antibody solution was aspirated and washed 10 times with PBST. Add 1 mL of 0.05 mol / L Gly-HCl solution to the immunotube and wait at room temperature for 5 minutes to elute the antigen-bound phages. Add 125 μL of 2 mol / L Tris solution to the ...
Embodiment 2
[0018] Example 2 Phage ELISA identification of scFv specificity
[0019] 30 clones were randomly selected from the plates after the fourth round of enrichment screening and cultured overnight. The next day, the strains were retained and re-activated by shaking. After the activated bacterial liquid grew to the logarithmic phase, the helper phage was infected, and the Kan resistance was supplemented overnight to induce the phage. The ELISA plate was coated with recombinant PSMA antigen overnight. The next day, the induced phage was used as the primary antibody, and the HRP-labeled anti-M13 antibody was used for ELISA detection.
[0020] Phage antibodies after the fourth round of enrichment were coated with PSMA recombinant protein on a 96-well plate, and the phage culture supernatant was screened by ELISA. A total of 30 clones were screened, and the positive standard was greater than 2 times the A value of the negative control. Among them, 3 clones were positive, and the posi...
Embodiment 3
[0021] Example 3 Determination of Nucleic Acid Sequence and Homology Analysis.
[0022] The above three positive clones were sequenced by Suzhou Jinweizhi Company after amplification of antibody variable region genes. Homology comparisons were performed using the Nucleotide Blast analysis software in NCBI. The results show:
[0023] The heavy chain nucleotide sequence is 94.68% homologous to the human immunoglobulin heavy chain variable region nucleotide sequence,
[0024]
[0025] The light chain nucleotide sequence is 97.22% homologous to the human immunoglobulin light chain nucleotide sequence.
[0026]
[0027]The sequencing results of the three positive clones showed that the nucleic acid sequences were basically identical. The nucleotide sequence encoding the heavy chain is shown in SEQ ID NO.1, and the nucleotide sequence encoding the light chain is shown in SEQ ID NO.2.
[0028] Homology comparisons were performed using the Nucleotide Blast analysis software ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com