Method for highly efficiently deleting large fragments of genome based on endogenous CRISPR-Cas system of zymomonas mobilis and application thereof
A technology of Zymomonas and deletion method, applied in the field of efficient deletion of large genome fragments, can solve the problems of host cell toxicity and difficult application, and achieve the effect of avoiding cytotoxicity and efficient deletion of large genome fragments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1 Construction of Zymomonas mobilis Endogenous CRISPR-Cas Genome Editing System
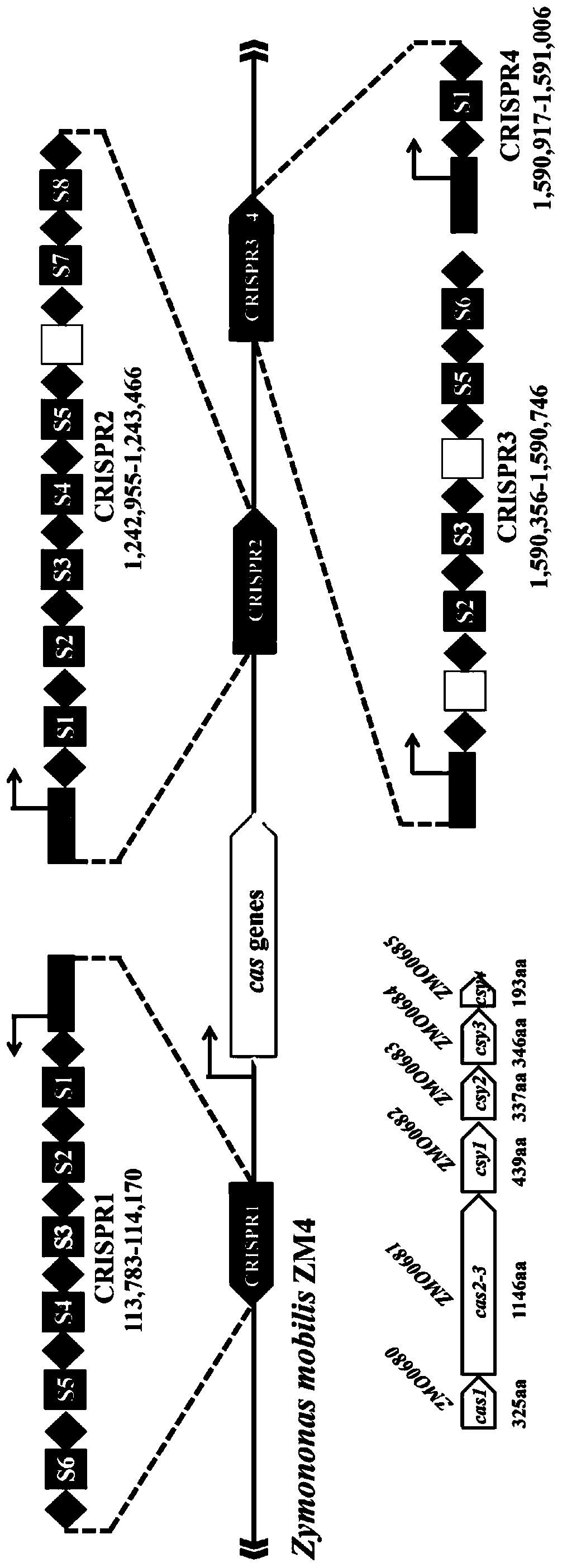
[0032] (1) CRISPR-Cas phylogenetic analysis of the genome encoding of Zymomonas mobilis
[0033] The premise of the present invention is that the research strain needs to encode the CRISPR-Cas system itself and have DNA splicing activity, which requires the host genome to encode the CRISPR cluster and the complete Cas protein system.
[0034] Taking Zymomonas mobilis Z.mobilis 4 as the model strain, the sequencing data of Z.mobilis 4 were analyzed. The results showed that the genome of the strain encoded four CRISPR structural sequences. According to their sequence in the genome, we sequenced them named CRISPR1-CRISPR4, see figure 1 . CRISPR1 occupies the 113,783-114,170 region of the genome and contains 7 repeat sequences; CRISPR2 occupies the 1,244,355-1,245,866 region and contains 9 repeat sequences; CRISPR3 occupies the 1,598,754-1,599,144 region and contains 7 repeat sequenc...
Embodiment 2
[0065] Example 2 Application of the method for efficient deletion of large genome fragments based on the endogenous CRISPR-Cas system of Zymomonas mobilis
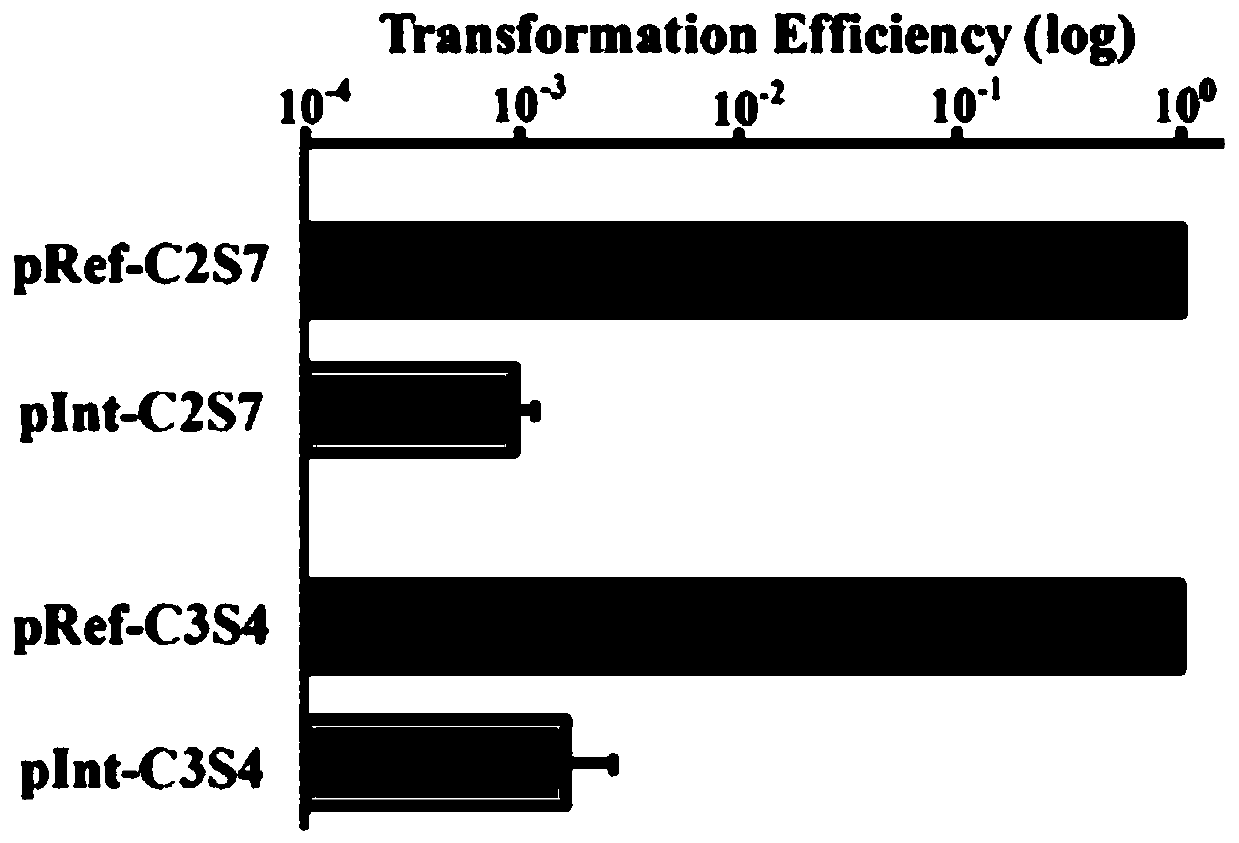
[0066] In the present invention, bioinformatics methods are firstly used to determine the essential genes that need to be retained and the non-essential genes that can be deleted, and select a non-essential gene with a length of 10 kb as the target knockout large fragment. Then design guide RNA and donor DNA sequences. Finally, the artificial CRISPR cluster expression module and donor DNA sequence were loaded on the plasmid, and the plasmid was electrotransformed into Zymomonas mobilis cells to complete the editing. Schematic diagram see Figure 5 As shown, the specific experimental scheme is as follows:
[0067] (1) Selection of target knockout large fragments
[0068] Through bioinformatics analysis, a non-essential gene ZMO1815-ZMO1822 (10,021bp) was found on the genome of the bacteria, which can be used as the targe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com