Double-plasmid system and application thereof
A plasmid and gene technology, applied in the field of dual-plasmid system for genome editing of Acinetobacter baumannii, can solve the genome editing problems of Acinetobacter baumannii
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1: pCasAb plasmid construction
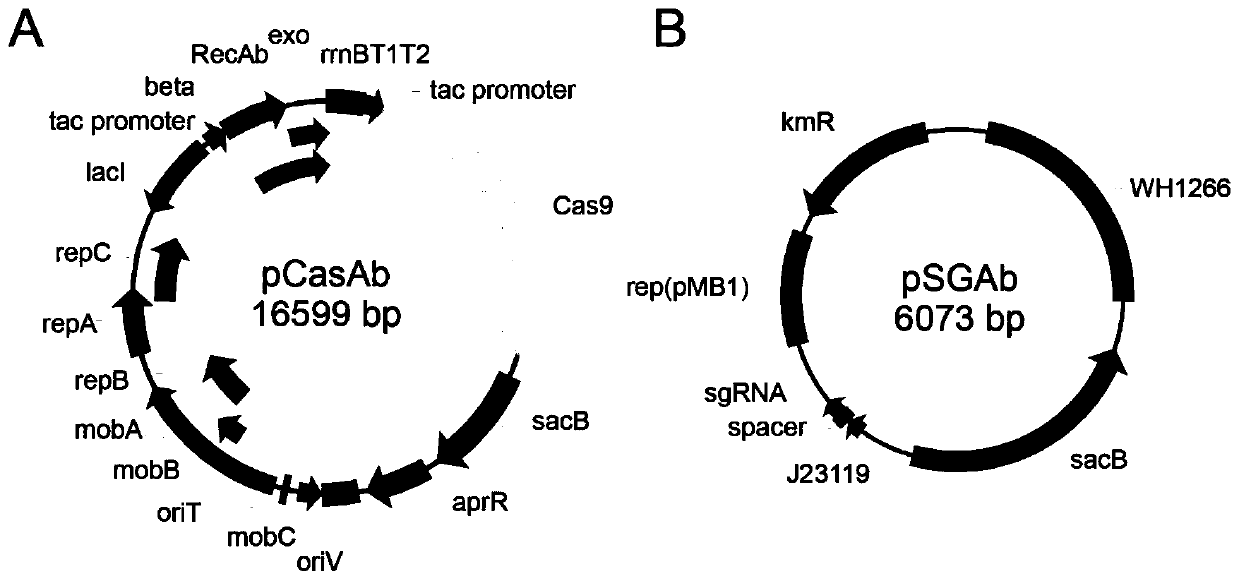
[0046] The composition of the pCasAb plasmid is attached figure 1 As shown in A, its sequence is SEQ ID NO: 1, and its specific construction method is as follows:
[0047] (1) The abramycin resistance gene fragment of aprR was amplified from the pCasKP-apr plasmid by polymerase chain reaction (PCR). The primer sequences used for PCR amplification are:
[0048] aprR-F: 5'-GCGTCAATTCACGGATCCGGTTCATGTGCAGCTCCATCAGC-3' (SEQ ID NO: 3)
[0049] aprR-R: 5'-AAACTTGGTCTGACAGTCAGCCAATCGACTGGCGA-3' (SEQ ID NO: 4)
[0050] Use Takara's PrimerSTAR HS DNA Polymerase to amplify the above DNA fragments, the reaction system is: 32 μL ddH 2 O, 4 μL dNTP Mixture (2.5mM each), 10 μL 5×Primestar Buffer, 1.5 μL aprR-F (10 μM), 1.5 μL aprR-R (10 μM), 0.5 μL pCasKP-apr plasmid template (1ng / μL), 0.5 μL PrimerSTAR HS DNA Polymerase.
[0051] After the system configuration is completed, carry out PCR amplification. Amplification parameters: pre-dena...
Embodiment 2
[0075] Embodiment two: pSGAb plasmid construction
[0076] The composition of the pSGAb plasmid is attached figure 1 As shown in B, its sequence is SEQ ID NO: 2, and its specific construction method is as follows:
[0077] (1) The WH1266 plasmid replicon DNA fragment was amplified from the pWH1266 plasmid by polymerase chain reaction (PCR). The primer sequences used for PCR amplification are:
[0078] WH1266-ori-F: 5'-TGAGAGTGCACCATAGGATTTTAACATTTTGCGTTGTTC-3' (SEQ ID NO: 9)
[0079] WH1266-ori-R: 5'-ATTTCACACCGCATAGCCAAGATCGTAGAAATATCTATGA-3' (SEQ ID NO: 10)
[0080] Use Takara's PrimerSTAR HS DNA Polymerase to amplify the above DNA fragments, the PCR reaction system is: 32 μL ddH 2 O, 4μL dNTP Mixture (2.5mM each), 10μL 5×Primestar Buffer, 1.5μL WH1266-ori-F (10μM), 1.5μL WH1266-ori-R (10μM), 0.5μL pWH1266 plasmid template (1ng / μL), 0.5 μL PrimerSTAR HS DNA Polymerase.
[0081] After the system configuration was completed, PCR amplification was carried out. The amplifi...
Embodiment 3
[0090] Example three: inserting the spacer fragment into the pSGAb plasmid
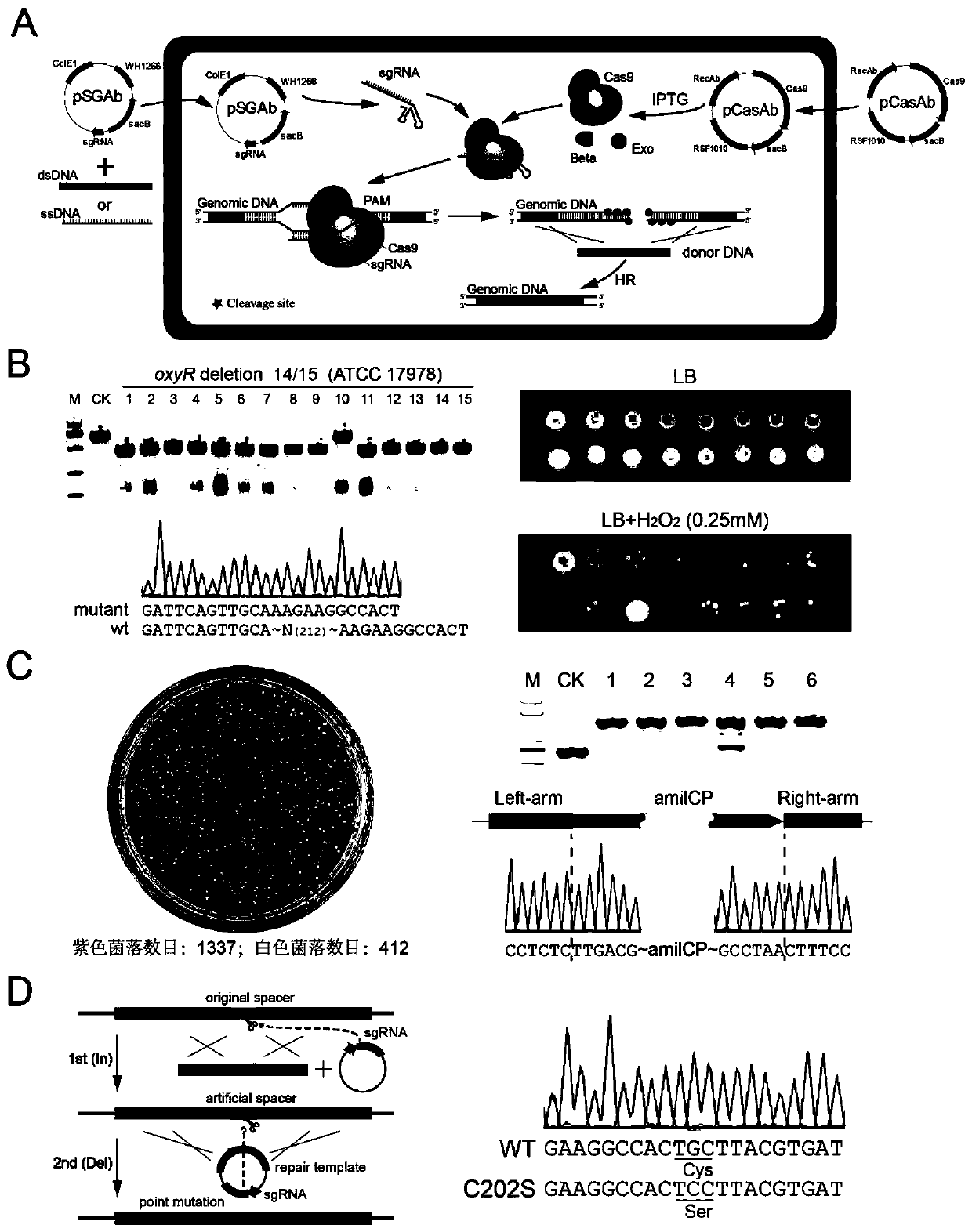
[0091] In order to enable the sgRNA and Cas9 nuclease complex to locate at a specific target site of Acinetobacter baumannii genomic DNA for cutting, the spacer sequence of the target site needs to be inserted into the sgRNA expression gene, and the insertion method is as follows:
[0092] (1) First, select the 20 base sequences before a certain NGG (N stands for any base of A / T / G / C) sequence on the target gene (these 20 base sequences are called spacers, and NGG is not included in Among them, the GC ratio of the spacer sequence is controlled at 20-80%). The feature of this step is that in order to insert the spacer fragment into the pSGAb plasmid, it is necessary to add a tagt adapter at the 5' end of the sense strand of the spacer sequence, and at the same time add an aaac adapter at the 5' end of the antisense strand of the spacer sequence. The specific primer design template as follows:
[0093]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com