Method for effectively screening membrane protein interacting protein based on yeast two-hybrid technology
A yeast two-hybrid, membrane protein technology, applied in the field of biomolecules, can solve problems such as affinity reduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0078] Take Populus trichocarpa Potri.001G190400.1 as an implementation case
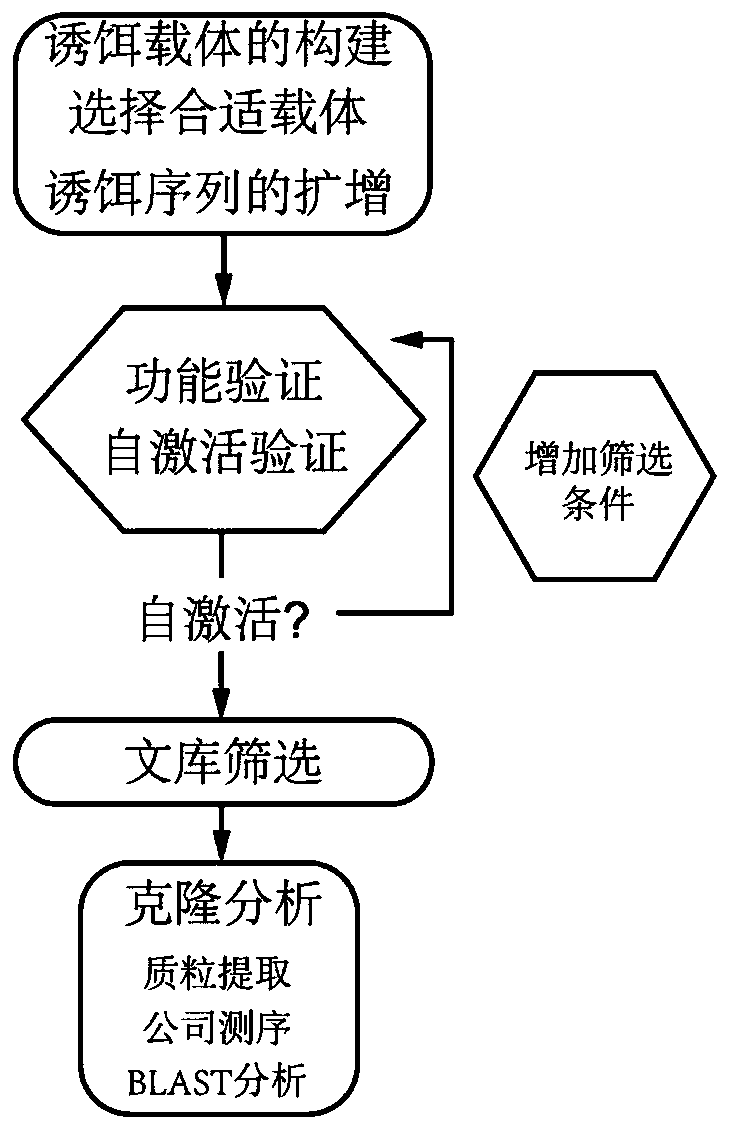
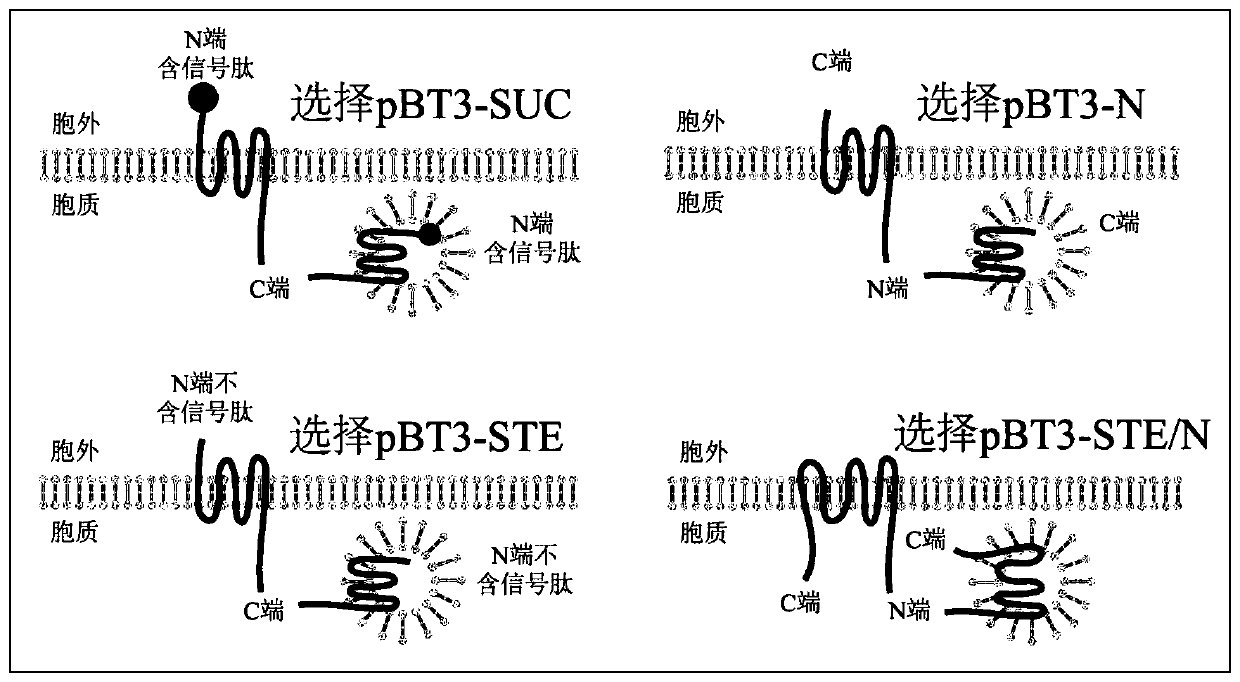
[0079] Step 1: Analyze the structure of the bait protein, design primers, and use Sfi1 enzyme as the restriction site to design primers to construct the bait vector.
[0080] pBT3-N:F ATTAACAAG GCC ATT ACG GCCATGGAGAAGAAGGTGGTGGT R TTA AAG ATACAG TGA ATG TGA GGCCGCCTCGGCCAATCAGTT
[0081] pBT3-SUC:F ATTAACAAG GCC ATT ACGGCCATGGAGAAGAAGGTGGTGGT RTTAAAGATACAGTGAATGTGAGGGGCCGCCTCGGCCAATCAGTT
[0082] pBT3-STE:F ATTAACAAG GCC ATT ACG GCCATGGAGAAGAAGGTGGTGGT
[0083] R TTAAAGATACAGTGAATGTGAGGGGCCGCCTCGGCCAATCAGTT
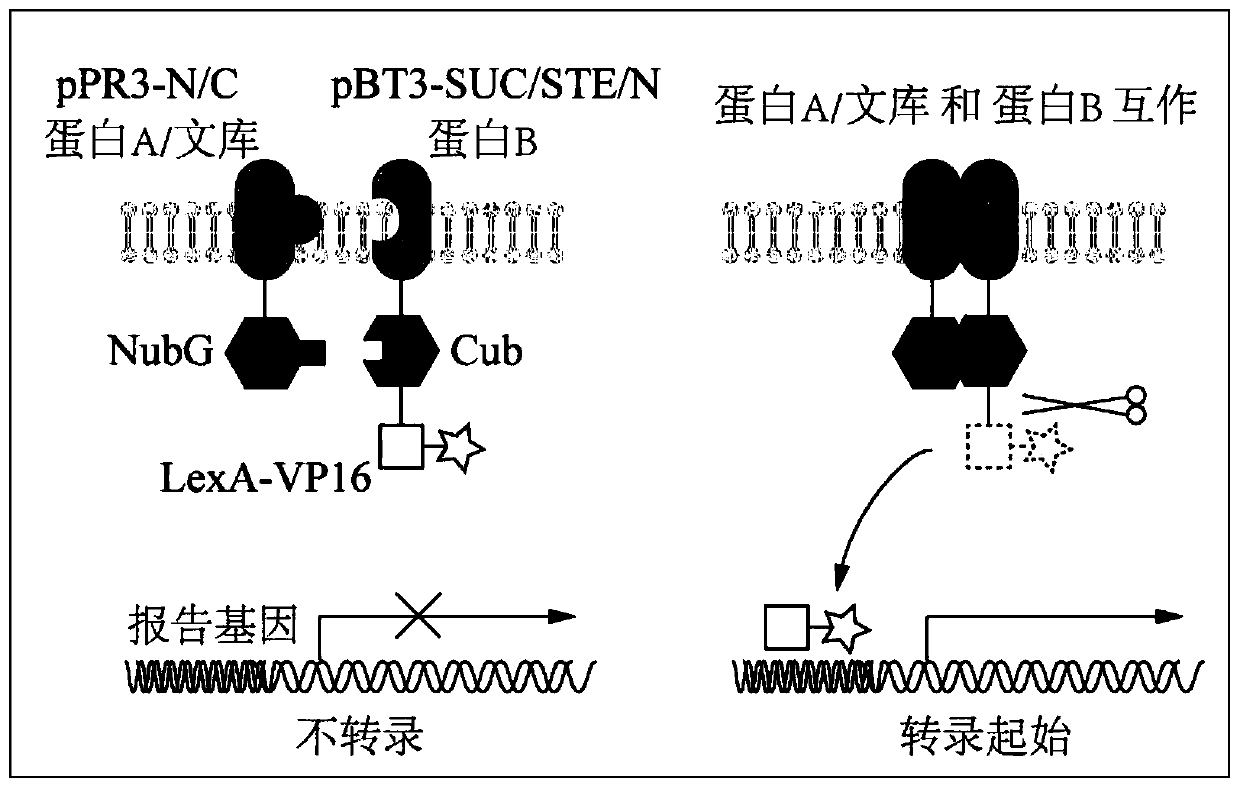
[0084] Step 2, self-activation detection and bait carrier function verification, using plasmids pTSU2-APP and pPR3-N as negative controls, and plasmids pTSU2-APP and pNubG-Fe65 as positive controls.
[0085] 1) Streak the NMY51 yeast strain on the YPDA plate, and culture it upside down at 30°C for about 2 days;
[0086] 2) Pick several yeast clones with a pipette tip and place them in 50ml...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com