Rat long-chain non-coding lncRNA-lncMSTRG10078 and application of rat long-chain non-coding lncRNA-lncMSTRG10078 in resisting cell damage
A long-chain non-coding and cell-damaging technology, applied in the field of molecular biology, can solve the problems of low lncRNA expression abundance, unconfirmed and undiscovered lncRNA, and achieve the effect of wide application value and resistance to damage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Embodiment 1 lncRNA-lncMSTRG10078 full-length sequence amplification
[0045] 1. Materials and Reagents
[0046] 1.1 Materials
[0047] The cell line used in this experiment is rat pituitary tumor cells (GH3 cells).
[0048] 1.2 Reagents
[0049] Phanta Max Super-Fidelity DNA Polymerase, Cat. No. P515, purchased from Nanjing Novizyme Biotechnology Co., Ltd.; HiScript II 1st Strand cDNA Synthesis Kit (+gDNA wiper), Cat. No. R212-01, bought from Nanjing Novozyme Biotech Co., Ltd. ; FastPure Gel DNA Extraction Mini Kit, item number DC301, purchased from Nanjing Nuoweizan Biotechnology Co., Ltd.; Mighty TA-cloning Reagent Set for Product No. 6019 was purchased from Bio-Technology (Beijing) Co., Ltd. (takara China); primers were synthesized by Nanjing GenScript Biotechnology Co., Ltd.
[0050] 2. Experimental method
[0051] 2.1 Extraction of total RNA
[0052] Total RNA was isolated from each GH3 cell sample in strict accordance with Sangon Bioengineering (Shanghai) ...
Embodiment 2
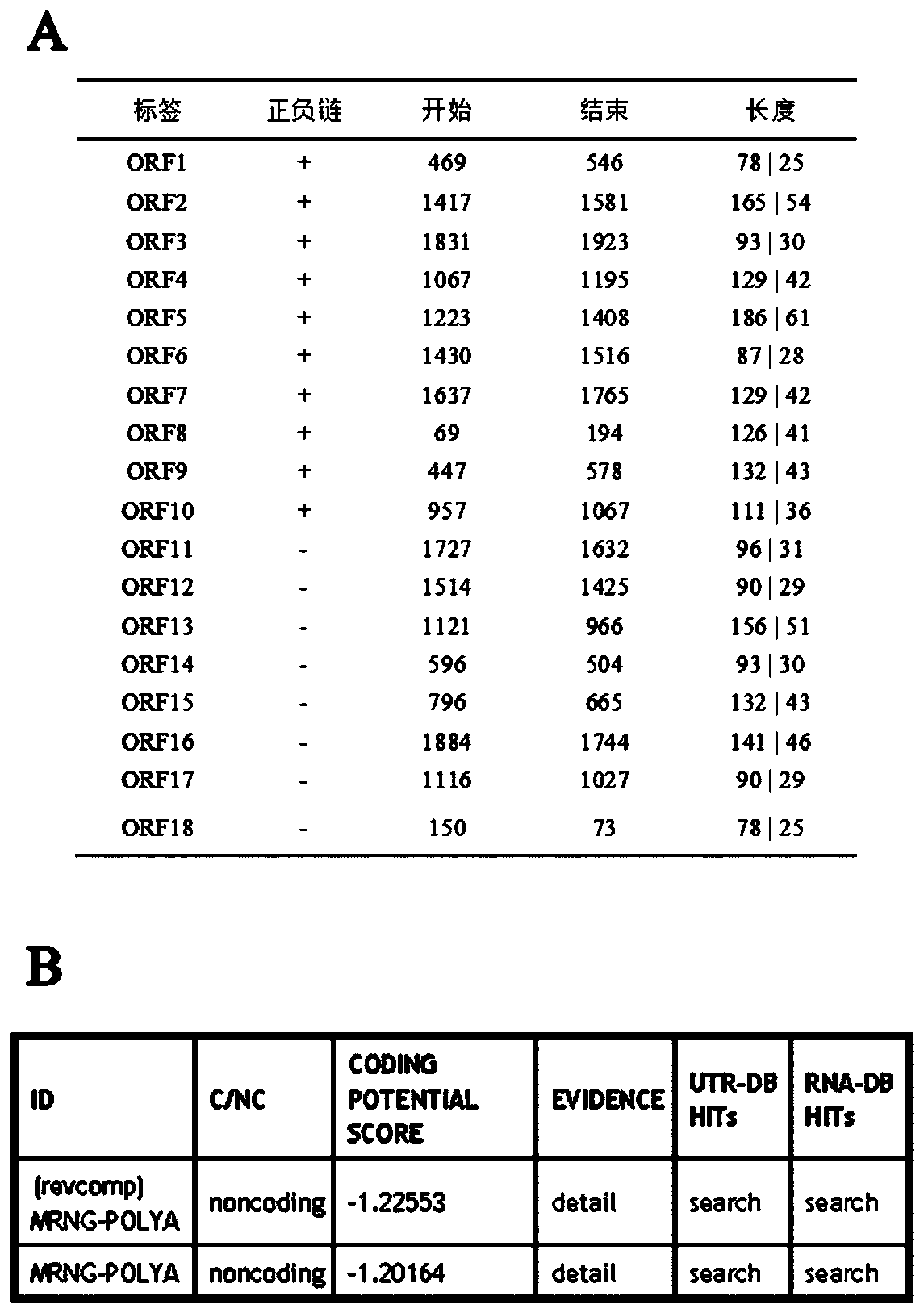
[0066] The full length of lncRNA-lncMSTRG10078 was identified using 5' and 3' RACE technology, and its full-length sequence was finally obtained as 1964bp, as shown in SEQ ID NO.1. Using UCSC and NCBI for sequence comparison, the results showed that lncRNA-lncMSTRG10078 was mainly located on chromosome 1 of rats (antisense strand, from 153859895-153861846) ( figure 1 ). Apply NCBI ORF-Finder and CPC, figure 2 It can be concluded that lncRNA-lncMSTRG10078 does not have protein coding ability, and the above results prove that lncRNA-lncMSTRG10078 is an lncRNA with a full length of 1964bp and no protein coding ability. Cloning and Analysis of Example 2 lncRNA-lncMSTRG10078 Sequence
[0067] 1. Reagents and carriers
[0068] Phanta Max Super-Fidelity DNA Polymerase, Cat. No. P505, purchased from Nanjing Novizyme Biotechnology Co., Ltd.; HiScript II 1st Strand cDNA Synthesis Kit (+gDNA wiper), Cat. No. R212-01, bought from Nanjing Novozyme Biotech Co., Ltd. ; FastPure Gel DNA ...
Embodiment 3
[0083] Example 3: Functional verification of lncRNA-lncMSTRG10078
[0084] 1. GH3 cell culture
[0085] GH3 cells were recovered in 5 mL DMEM complete medium (10% FBS + 90% DMEM + 1 mL double antibody + 1 mL glutamine) and placed in an incubator with 95% relative humidity at 37°C and 5% CO2 nourish. When the growth state of the cells reaches 70% to 90%, the trypsin-EDTA digestion method is used to pass passage at a ratio of 1:2, and pass passage once every 1-2 days. Cell cryopreservation adopts cell cryopreservation medium (90% FBS+10% DMSO) to carry out cryopreservation.
[0086] 2. Exploration of GH3 cell transfection conditions
[0087] The day before transfection, trypsinize the cells and count (it is advisable to make the density reach about 90% before transfection after about 24 hours), and plate the cells in 1 mL of normal growth medium containing serum and without antibiotics; Dilute 1:1, 1:1.5, 1:2, 1:2.5, 1:3 ExFect Transfection Reagent with 50 μL of serum-free O...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com