Identification method of group of genomic characteristic mutation fingerprints associated with homologous recombination repair defects
A homologous recombination, genome technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve problems such as statistical differences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
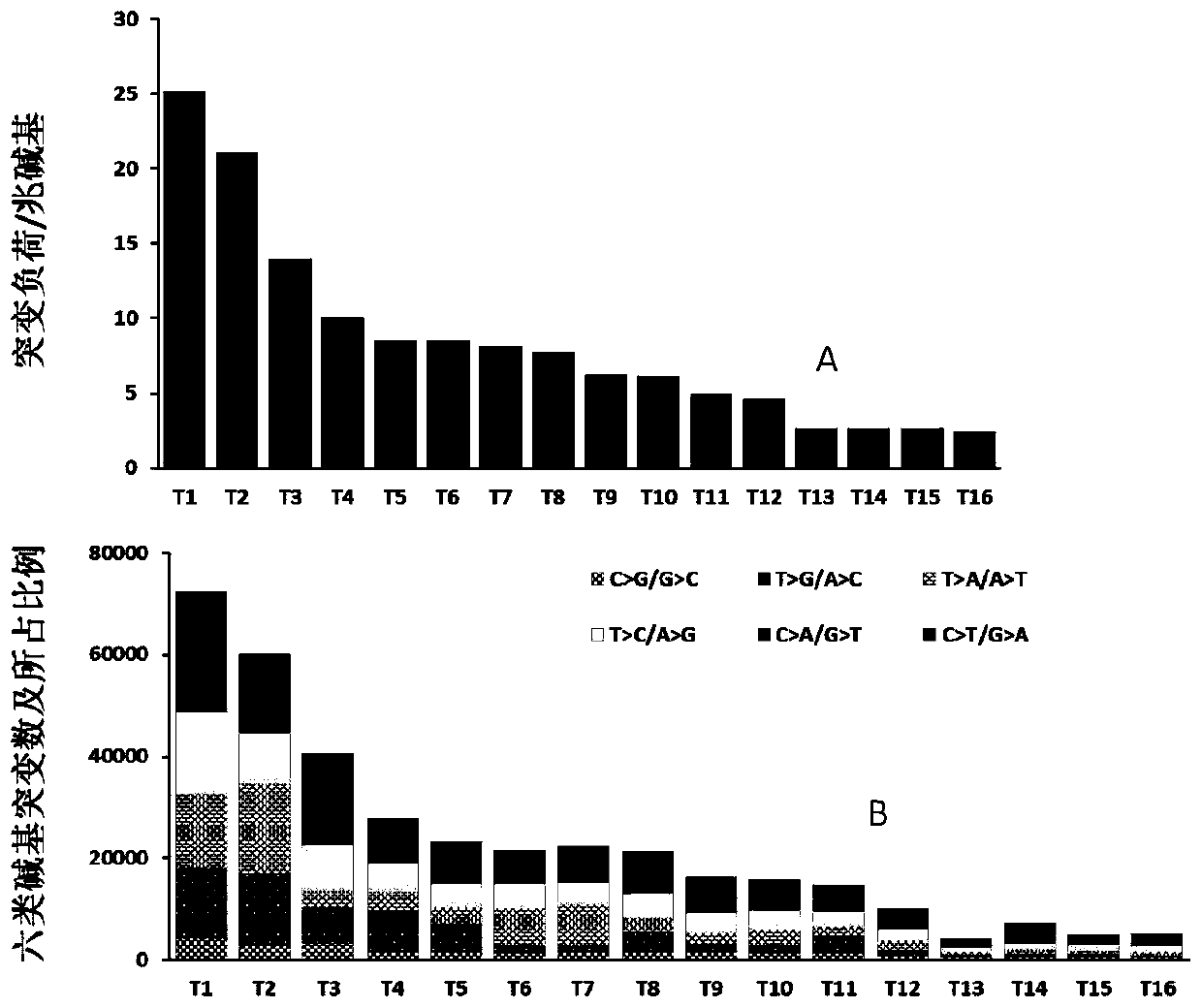
[0059] Example 1 Genomic Tumor Mutation Burden of HBV-related Liver Cancer
[0060] Methods: Surgical sampling of fresh liver cancer and blood tissue, sampling volume> 1cm 3 , the blood sample was 5 ml of anticoagulated blood, and the blood cells were collected by centrifugation. The samples were quickly frozen in liquid nitrogen and placed in individual sealed packages to prevent contamination between samples, and basic clinical information was marked on the packaging containers. The samples were stored at -80°C and shipped to the sequencing company within two weeks on dry ice for sample quality inspection, nucleic acid extraction and sequencing. After genome purification, use Qubit and 1% AGE to detect the degradation of the sample. The concentration is ≥20ng / ul and the total amount is ≥500ng. The sample has no obvious degradation band and can be used for downstream analysis and sequencing.
[0061] The sequencing platform uses the illumina2000 sequencer, and the sequencin...
Embodiment 2
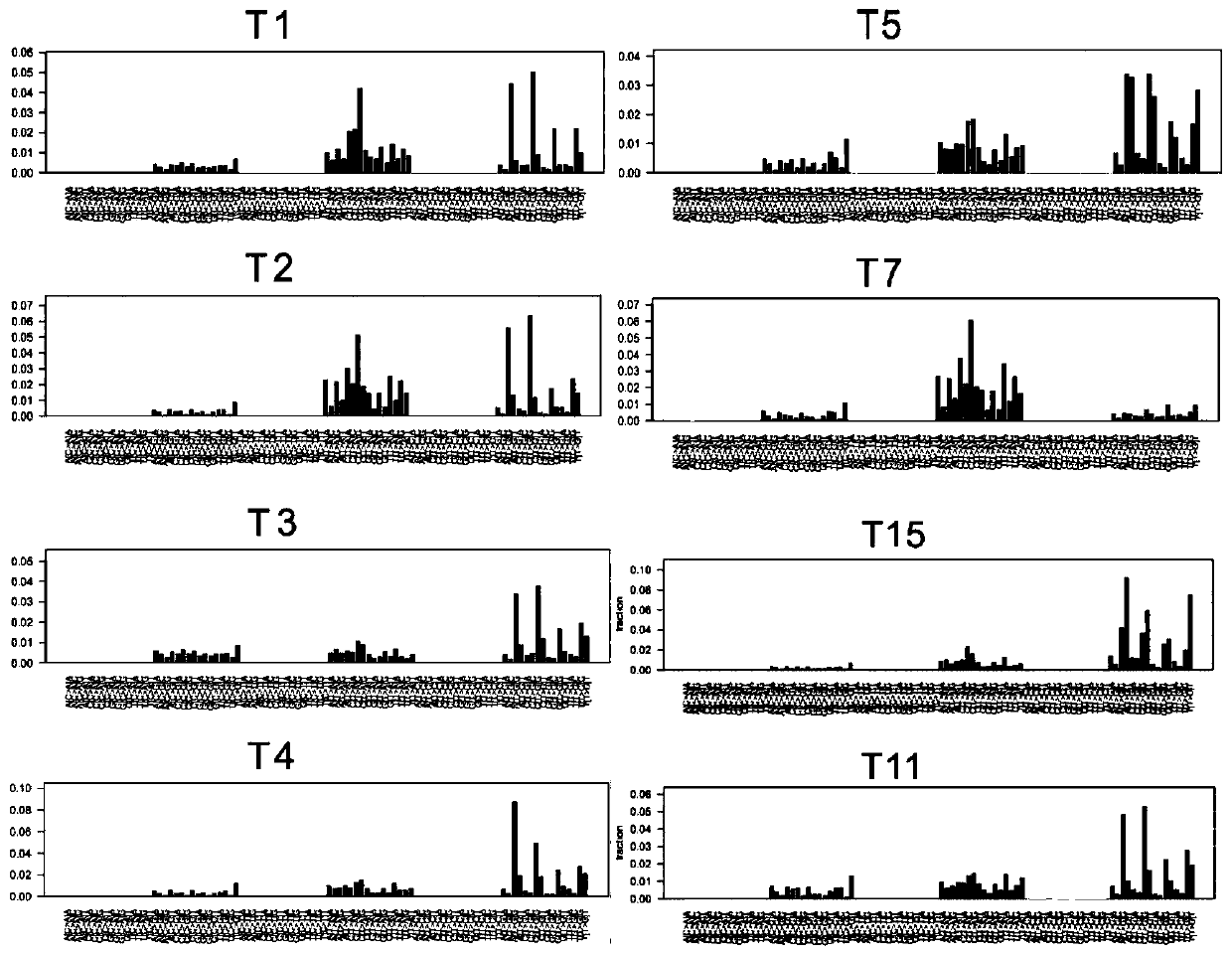
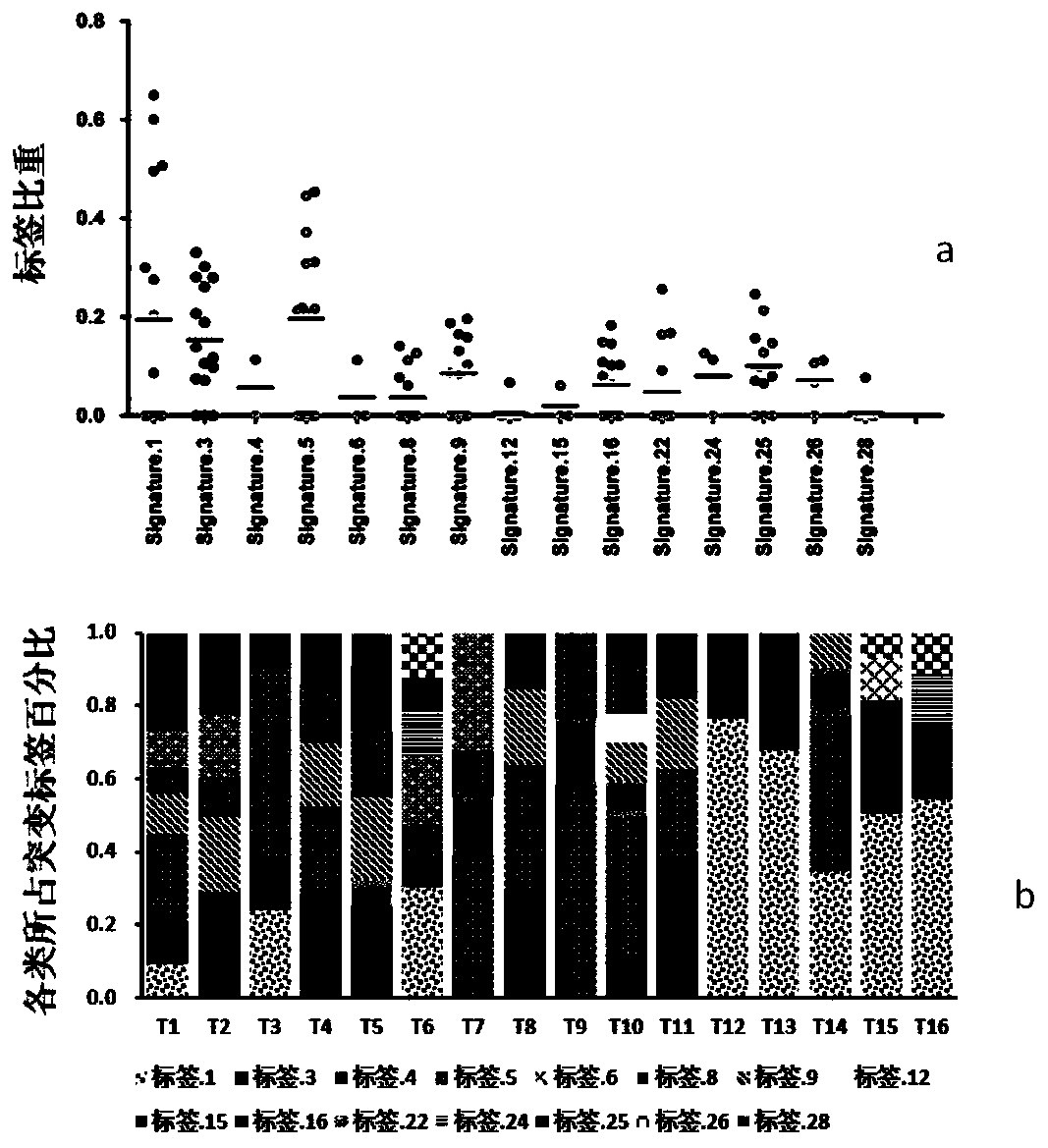
[0068] Example 2 Single nucleotide variation characteristics of hHRD type in HBV-related liver cancer genome
[0069] Method: According to the definition of 30 types of point mutation signatures published by the Sanger Institute in Cambridge, UK in 2013 (Nature.2013 Aug22; 500 (7463): 415-21), the 16 cases of HBV in Example 1 were further correlated Analyze the clustering characteristics of single nucleotide variants in liver cancer samples. The single-nucleotide variation label extraction strategy adopts the 30 types of standard single-nucleotide variation characteristics defined in the COSMIC database in a single sample, and uses the deconstructSigs software to decompose the mutation characteristics and their composition ratios in a single tumor sample (
[0070] Genome Biol. 2016 Feb22;17:31., the full text is incorporated by reference). The deconstructSigs software analysis method can perform single-nucleotide variation tag analysis on a single sample. Compared with the...
example 3
[0095] Example 3 Compositional characteristics of genome-wide small fragment insertion-deletion mutations in HBV-related liver cancer
[0096] Method: The specific analysis method of small fragment indels is as follows: Figure 8 show:
[0097] Small fragment indel detection software uses Strelka. The analysis strategy or method for small fragment insertion-deletion features is
[0098] 1. The small fragment deletion events that define the repeat sequence are 1-50bp small fragment deletions that occur in the short tandem repeat region.
[0099] 2. Define the small fragment deletion event of the micro-homology sequence as the 1-50bp small fragment deletion in the non-short tandem repeat region, accompanied by homology sequences > 3bp on both sides.
[0100] 3. Define other types of deletions as small fragment deletions of 1-50 bp in non-short tandem repeat regions, and not accompanied by homologous sequences > 3 bp on both sides, and the range of homologous sequences is defi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com