Method for improving production capacity of L-arginine strain
A production capacity, arginine technology, applied in the field of genetic engineering, can solve the problems of low fermentation level, weakened branched metabolic pathways, etc., and achieve the effect of improving production capacity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] Example 1: Preparation of strain ATCC13032 (ΔargR) with argR gene knockout
[0082] 1.1 ATCC13032 genome extraction: connect a small amount of ATCC13032 glycerol bacteria (purchased from Shanghai Institute of Industrial Microbiology) to the BHIS test tube, culture on a constant temperature shaker at 30°C, 220rpm for 18 hours, collect the bacteria by centrifugation at 12000rpm, and use Axygen bacterial genome small-scale extraction reagent Cassette extraction of the ATCC13032 genome.
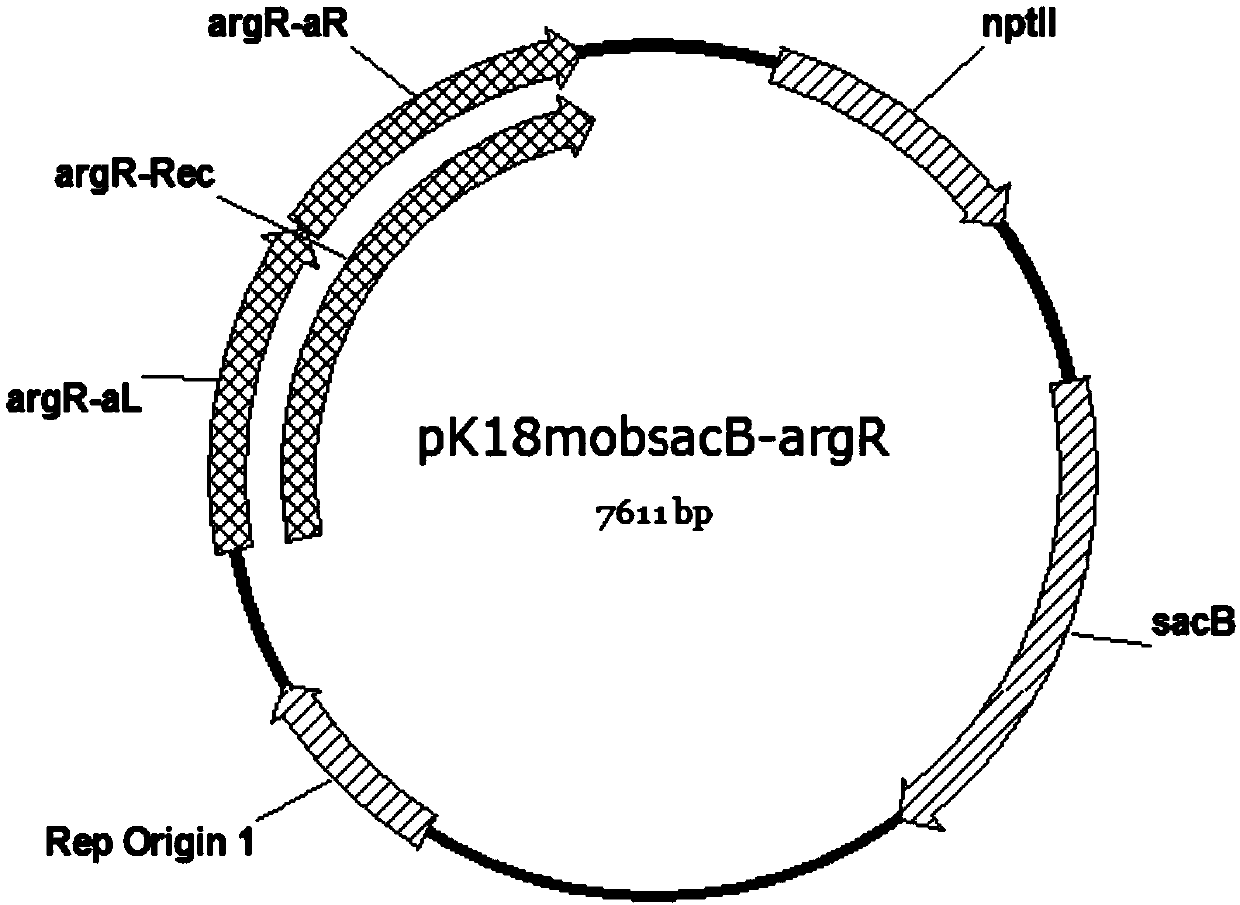
[0083] 1.2 Construction of knockout plasmid pK18mobsacB-argR
[0084] 1.2.1 Using the ATCC13032 genome extracted in step 1.1 as a template, use primers argR-aL-F and argR-aL-R to perform PCR amplification of the argR-aL fragment. The PCR system is as follows (the following PCR reagents were purchased from Toyobo TOYOBO KOD series): KOD Buffer 5μl, dNTP 5μl, MgSO 4 4 μl, primer argR-aL-F 0.5 μl, primer argR-aL-R 0.5 μl, KOD Plus Neo 1 μl, template 0.4 μl, ddH 2 O to make up 50 μl. The ...
Embodiment 2
[0095] Example 2: Preparation of strain ATCC13032 (ΔargRargBmut (A26V M31V) with mutant argB gene
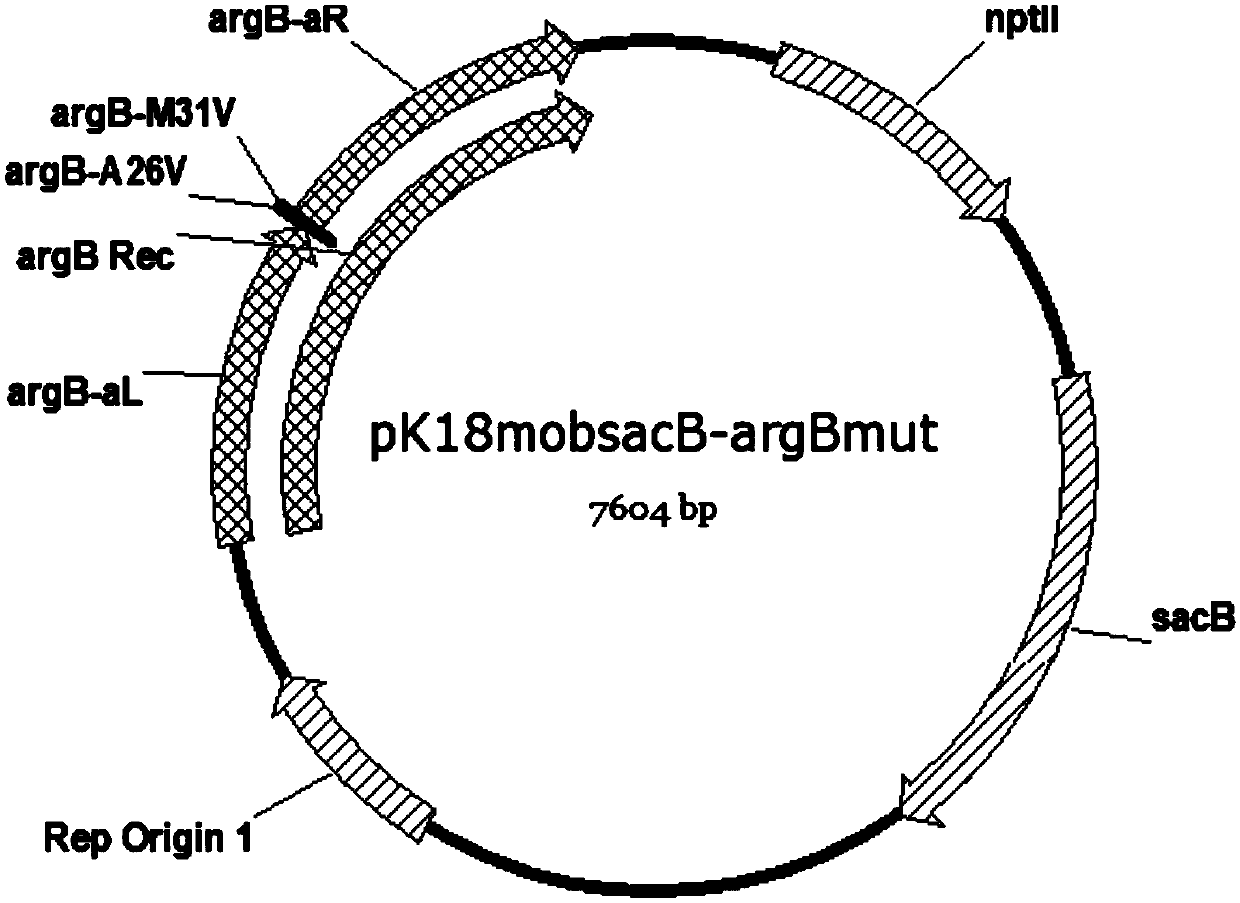
[0096] 2.1 Construction of mutant plasmid pK18mobsacB-argBmut:
[0097] 2.1.1 Using the ATCC13032 genome extracted in step 1.1 as a template, use the primers argB-aL-F and argB-aL-R to amplify the argB-aL fragment according to the same PCR conditions as step 1.2.1, about 1kb, for the purpose of gel recovery fragment.
[0098] 2.1.2 Using the ATCC13032 genome extracted in step 1.1 as a template, use the primers argB-aR-F and argB-aR-R to amplify the argB-aR fragment according to the same PCR conditions as step 1.2.1, about 1kb, for the purpose of gel recovery fragment.
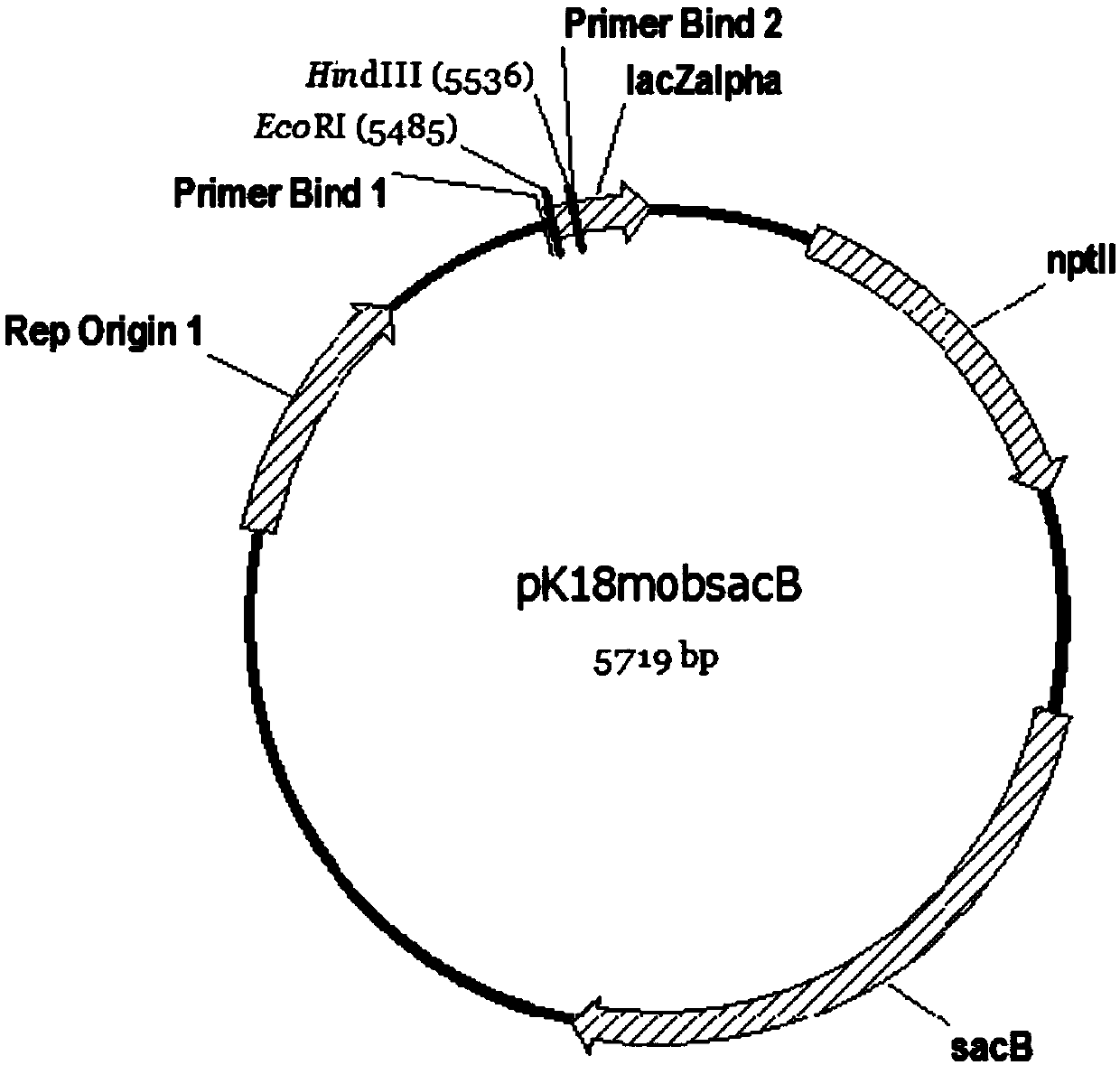
[0099] 2.1.3 Use Axygen's plasmid extraction kit to extract plasmid pK18mobsacB (gifted by Liu Shuangjiang, Institute of Microbiology, Chinese Academy of Sciences, its structure is as follows figure 1 shown), the plasmid was digested with HindIII and EcoRI, and the gel was recovered to obtain a 5.7kb vector fr...
Embodiment 3
[0109] Embodiment 3: the preparation of the bacterial strain CIBTS1512 (NCgl2620mut6) of the nucleotide sequence of the upstream regulatory region of the start codon of the mutation polyphosphate kinase gene (NCgl2620)
[0110] 3.1: Construction of recombinant plasmid pK18mobsacB-NCgl2620mut6
[0111] 3.1.1 Using the ATCC13032 genome extracted in step 1.1 as a template, use primers NCgl2620mut6-aL-F and NCgl2620mut6-aL-R to amplify the NCgl2620mut6-aL fragment, about 0.5kb, under the same PCR conditions as step 1.2.1, and recover from the gel target fragment.
[0112] 3.1.2 Using the ATCC13032 genome extracted in step 1.1 as a template, use primers NCgl2620mut6-aR-F and NCgl2620mut6-aR-R to amplify the NCgl2620mut6-aR fragment, about 0.5kb, according to the same PCR conditions as step 1.2.1, and recover from the gel target fragment.
[0113] 3.1.3 Use Axygen's plasmid extraction kit to extract plasmid pK18mobsacB (gifted by Liu Shuangjiang, Institute of Microbiology, Chinese...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com