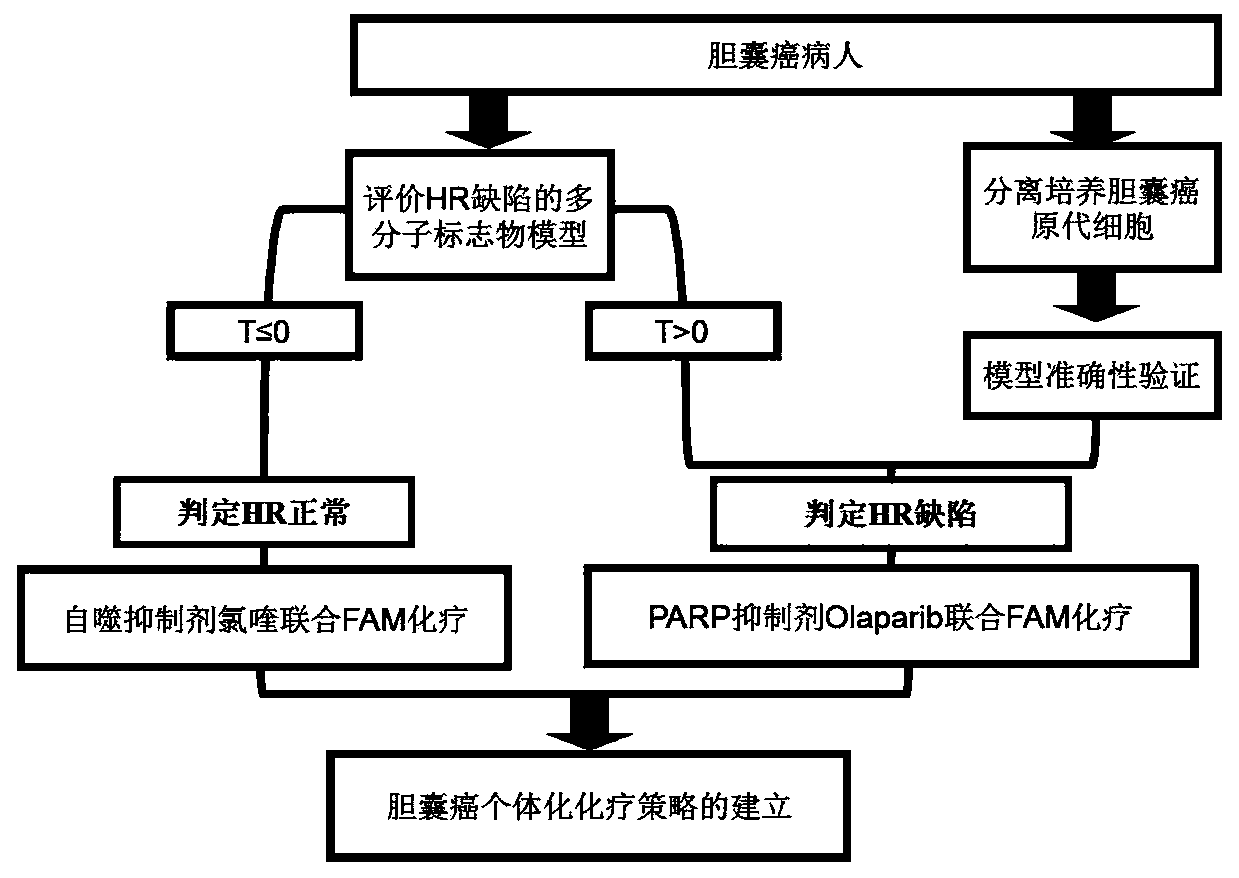
Model for evaluating HR defect and application
A defect model and defect technology, applied in the evaluation of multi-molecular markers of homologous recombination defects, evaluation of HR defect models and application fields, can solve the problem that the evaluation system of multi-molecular markers has not yet been found, and the combination drug or sequential drug has not been found. Drug administration and other issues, to achieve the effect of improving anti-tumor efficacy, easy clinical test detection, and convenient use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1: Construction of Multimolecular Markers for Evaluation of Homologous Recombination Deficiency and HR Deficiency Model
[0044] 1. Assessing the Selection of Multimolecular Markers in the HR Deficiency Model
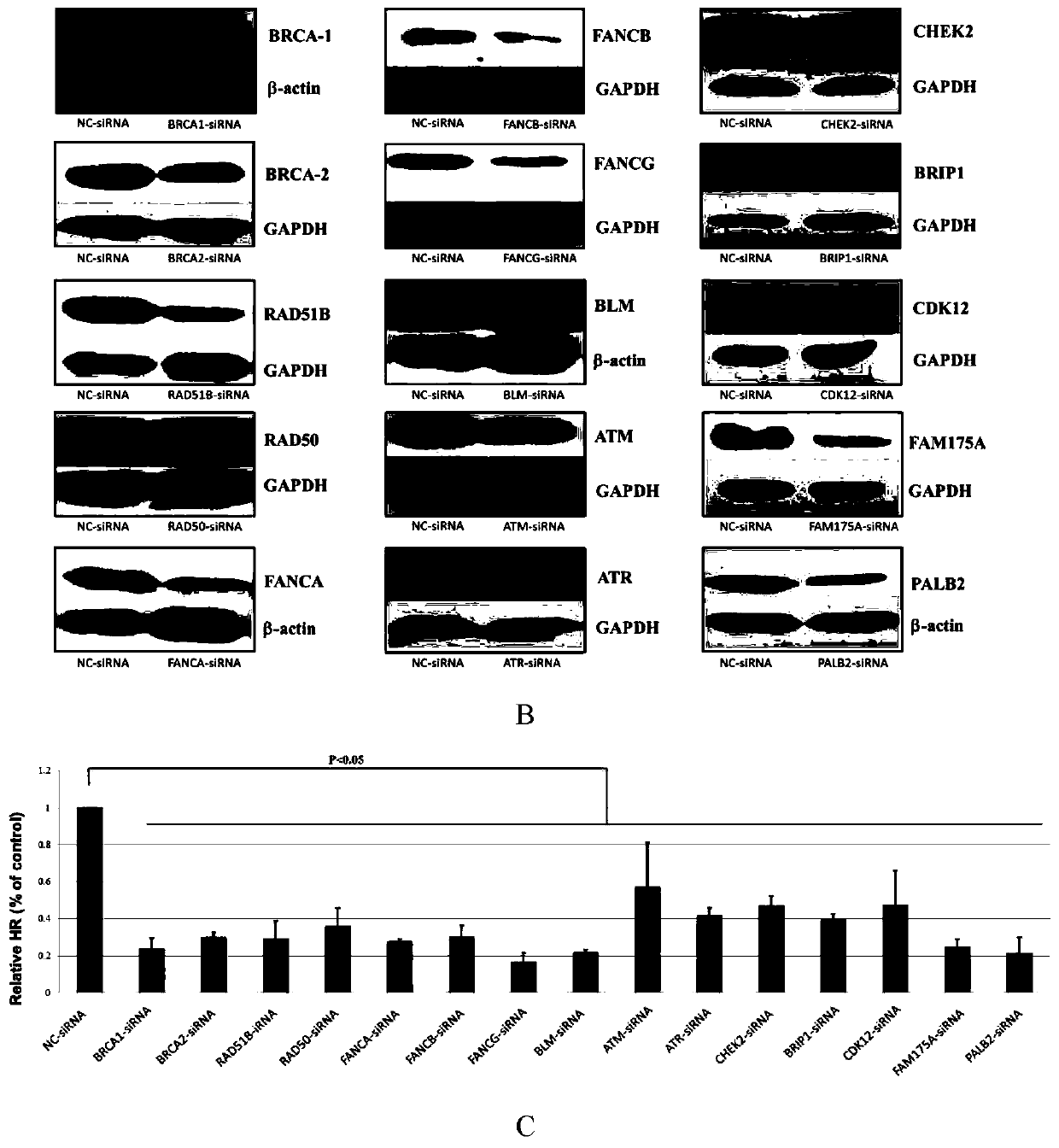
[0045] The DNA sequencing data of 553 gallbladder cancer patient samples were obtained from the database Catalog Of Somatic Mutations In Cancer (COSMIC, cancer.sanger.ac.uk / cancergenome / projects / cosmic), and the mutated genes were sorted according to the mutation frequency. Research on molecules related to the repair mechanism Selected 15 genes (M1-M15) that played a key role in them as multi-molecular markers for chemotherapy of gallbladder cancer targeting homologous recombination, as shown in Table 1.
[0046] Table 1. Multimolecular marker combinations for judging HR deficiency
[0047]
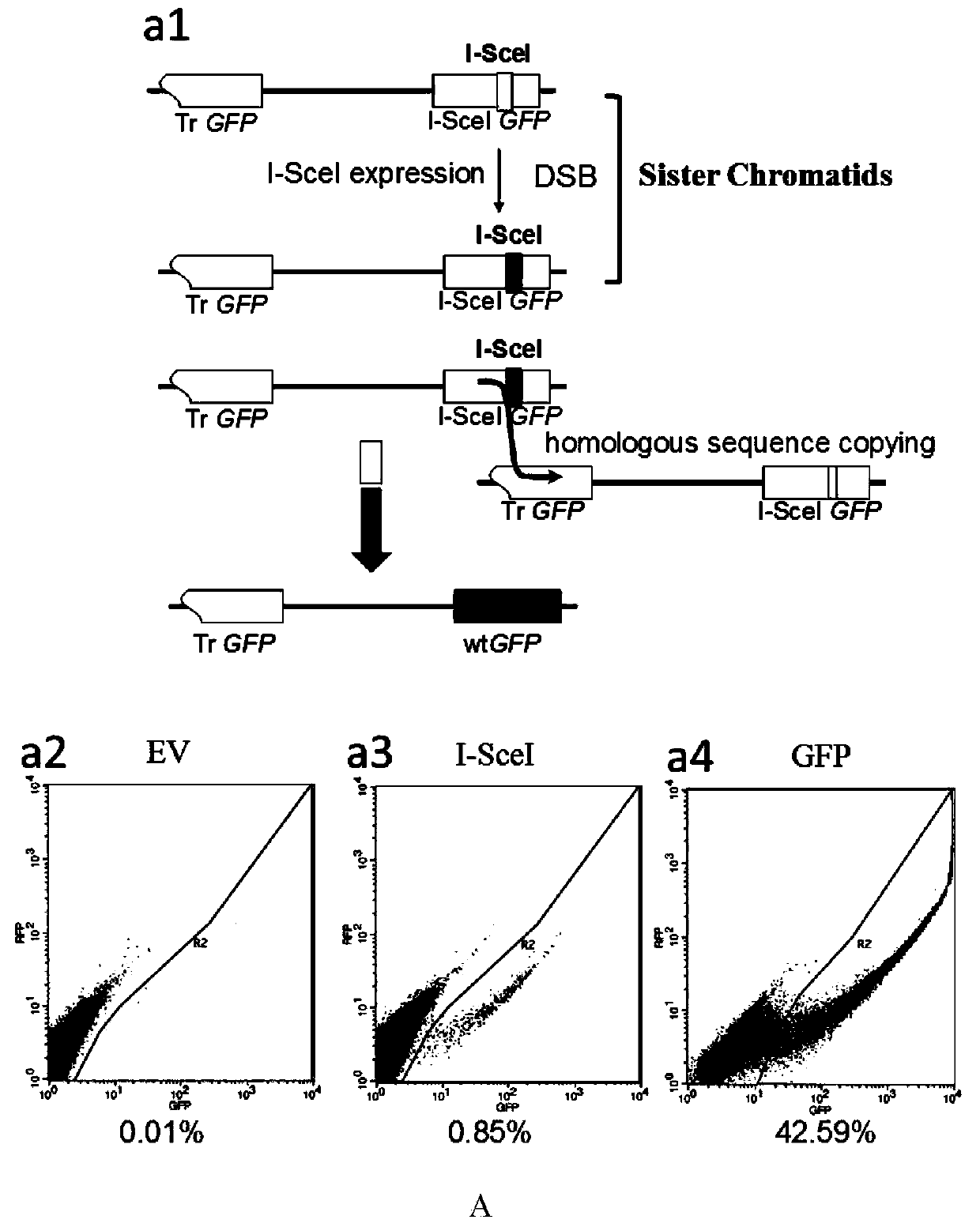
[0048] 2. Assessing Validation of Multimolecular Markers in HR Deficiency Models
[0049] 1) Puromycin concentration selection:
[0050] Gallbladder cancer GBC...
Embodiment 2
[0075] Example 2: Isolation and culture of primary gallbladder cancer cells and verification of the accuracy of the HR-deficient model
[0076] 1. Construction of gallbladder cancer GBC-SD and SGC-996 cell lines with stable knockdown of BRCA1 and BRCA2
[0077] (1) Lentivirus pre-infection experiment
[0078] Follow 3-5 x 10 3 GBC-SD and SGC-996 gallbladder cancer cells were pre-seeded in the wells of 96-well culture plate per well (1640 medium containing 10% FBS, volume 100 μl), and the cell fusion degree should be 30% during lentivirus infection -50% or so. 10 μl of virus solution (BRCA1, BRCA2 lentiviral particles are entrusted to Shanghai Jikai Biological Co., Ltd. to make the coating) according to the final concentration of 1×10 6 TU / ml, 1×10 5 TU / ml, 1×10 4 TU / ml 3 concentrations pre-infected cells. For example, the number of gallbladder cancer cells is about 1×10 4 1, the MOI of the above three wells is about 100, 10, 1. After culturing at 37°C for about 12 hours...
Embodiment 3
[0090] Example 3: The autotropic inhibitor chloroquine significantly enhances the killing effect of FAM on gallbladder cancer cells with normal HR
[0091] 1. Configure chloroquine:
[0092] An appropriate amount of chloroquine (Sigma, product number: C6628) was dissolved in double-distilled water to form a 10 mM stock solution, which was sterilized with a 0.22 μm filter and stored at 4°C. For in vitro cell experiments, it was diluted to 100 μM with 1640 medium for experiments. In vivo animal experiments were injected intraperitoneally according to the standard of 31.25ul / 10g at a concentration of 10mM.
[0093] The gallbladder cancer cells GBC-SD and SGC-996 used in this example were judged by the HR deficiency model described in Example 1 to have a T value of 0, that is, the HR was normal.
[0094] 2.5-FU treatment induces autophagy in gallbladder cancer cells:
[0095] (1) Gallbladder cancer cells GBC-SD and SGC-996 were treated with 5 μM 5-FU for 24 and 48 hours, respect...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com