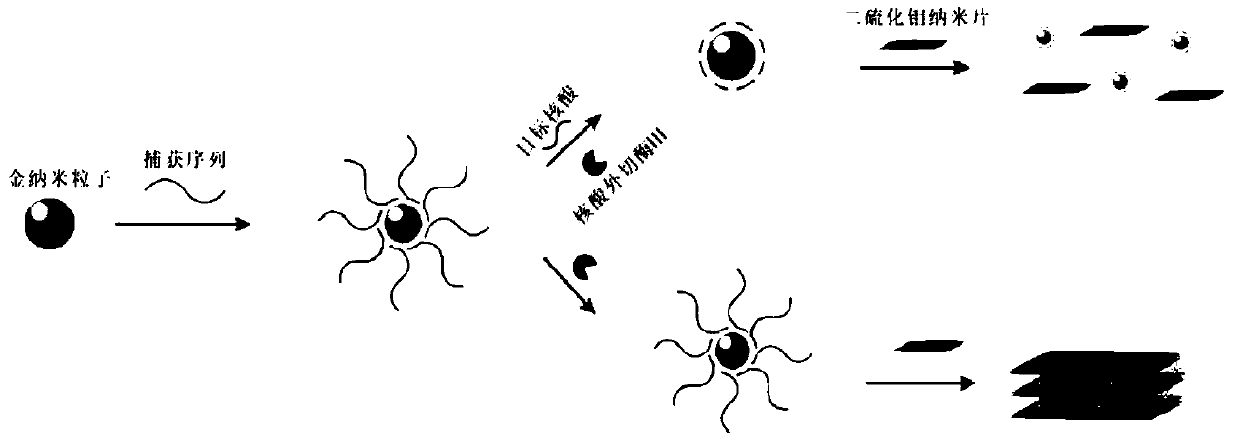
Method for visually detecting nucleic acid based on enzyme catalysis circulation and molybdenum disulfide adsorption mediation
A molybdenum disulfide and nucleic acid technology, applied in the field of colorimetric reagent research and nucleic acid detection, can solve the problems of complex operation and low sensitivity, and achieve the effects of good selectivity, improved detection sensitivity and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Nanogold-capture probe preparation:
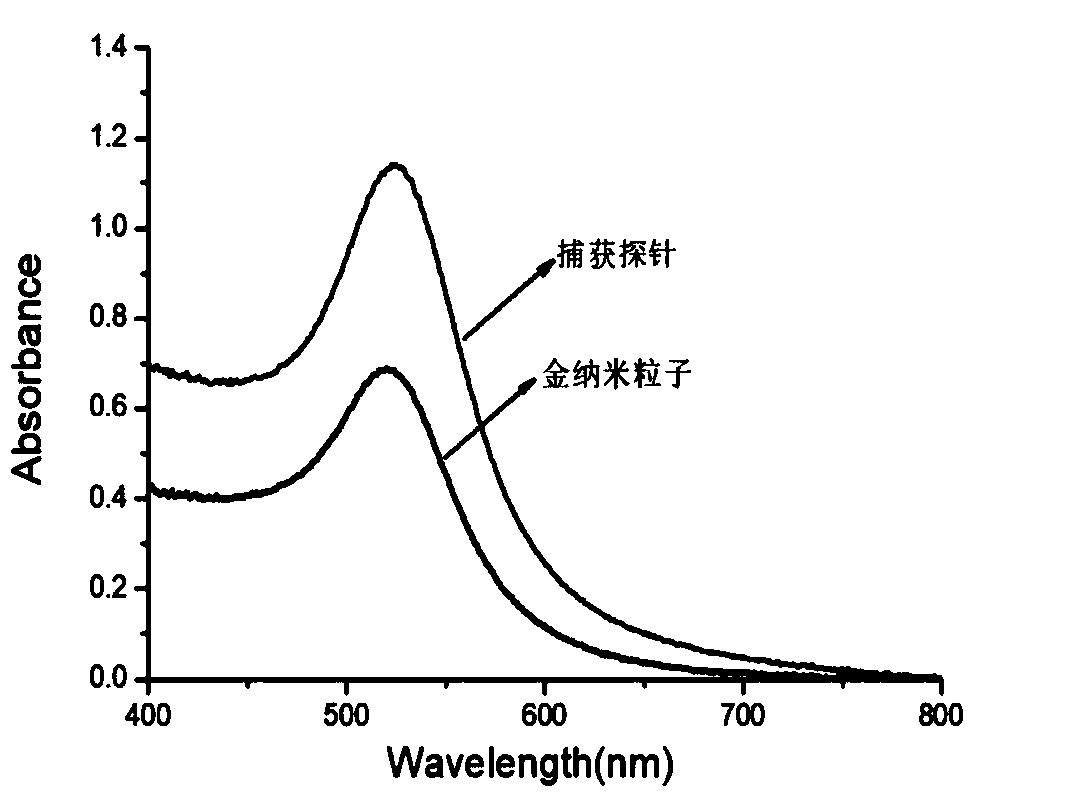
[0040] (1) Mix the polyA-modified target nucleic acid capture sequence with 15nm gold nanoparticles at a molar ratio of 200:1 (such as a final concentration of 600nM DNA: 3nM gold nanoparticles), and shake gently overnight at room temperature at 25°C. The capture sequence is: SEQ ID NO.15'-AAAAAAAAAAAAAAAAAAAAACTCTCTAAAATCACT-3'(capture sequence modified by polyA); in the above steps, the target nucleic acid capture sequence modified by sulfhydryl group can also be used, SEQ ID NO.2: 5'-SH-CTCTCTAAAATCACT-3'(capture sequence modified by sulfhydryl group capture sequence), the follow-up effect is consistent with the polyA modified target nucleic acid capture sequence.
[0041] (2) Add 1M PBS (100mM PB, 1M NaCl, pH=7.4) to the solution of step (1) by volume ratio, add once every 30min, and add in five times, so that the final concentration of the mixed solution is 0.1M PBS ( 10 mM PB, 0.1 M NaCl, pH=7.4), and continue shaking overnig...
Embodiment 2
[0046] Take 100 μL of the nano-gold capture probe solution prepared in Example 1 (the final concentration of the probe is 6 nM), and add the target nucleic acid with a final concentration of 200 nM to it respectively. The sequence of the target nucleic acid is: SEQ ID NO.3: 5' -AGTGATTTTAGAGAGAG-3', after hybridization at room temperature for 10 minutes, exonuclease III at a final concentration of 0.1 U / μL was added, and reacted at 37°C for 30 minutes. Then add molybdenum disulfide nanosheets (size 200nm×200nm) with a final concentration of 15 μg / mL, and the transmission electron microscope image of molybdenum disulfide is shown in figure 2 ), let stand to react for 70min, take the supernatant to observe the color change of the supernatant with naked eyes, and measure the absorbance value change of the supernatant by an ultraviolet spectrophotometer.
Embodiment 3
[0048] The method of embodiment 3 is the same as that of embodiment 2, except that the particle diameter of gold nanoparticles is 10nm; the hybridization time between target nucleic acid and capture probe is 30min; the final concentration of exonuclease III is 0.05U / μL, The shearing time is 90min; the size of the molybdenum disulfide nanosheets is 100nm*100nm, and the final concentration is 18μg / mL; the static reaction time is 10min.
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com