A molecular marker locus linked to the content of tea plant epicatechin and its application
An epicatechin and molecular marker technology, applied in the field of molecular genetics and breeding, can solve problems such as failure to find and affect the content of epicatechin, and achieve the effect of great research value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] 1. Experimental samples
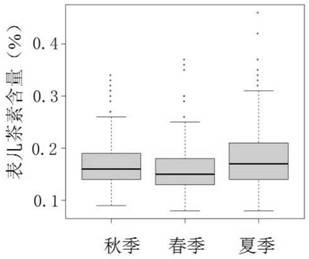
[0053] Collected 191 tea tree materials located in the Guangdong Tea Tree Germplasm Resource Bank (Guangdong, Yingde, 113.3OE, 24.3ON), including 124 in Guangdong, 20 in Fujian, 15 in Guangxi, 9 in Zhejiang, 6 in Hunan, and 6 in Yunnan. 1 copy in Jiangxi, 1 copy in Guizhou, and 1 copy in Taiwan. In addition, 8 descendants of Kenyan tea species and 1 descendant of Georgian species, the selected materials are widely representative.
[0054] The selected resources are randomly distributed in the resource pool. Single planting in double rows, each row 4m, row spacing 1.5m, plant spacing 35cm. The resource bank conducts routine water and fertilizer management. At the end of 2016, the resources were pruned and basal fertilizer was applied in deep pits, 4 tons of organic fertilizer, 0.75 tons of peanut bran and 10 catties of compound fertilizer per mu. After spring tea and summer tea in 2017, pruning and topdressing outside the roots, 30 catties o...
Embodiment 2
[0079] Example 2 Verification of molecular markers in another population
[0080] 1. Experimental method
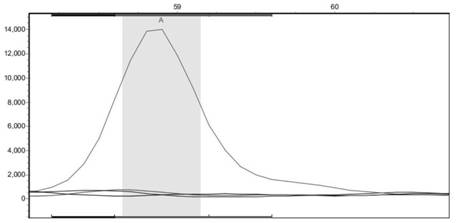
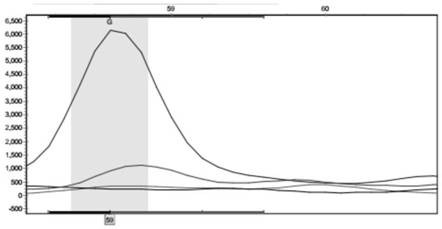
[0081] The SNP site located at Scaffold2233:468642 was verified in another population containing 98 germplasm.
[0082] 1. Detect the epicatechin content of each sample. The specific detection method is the same as in Example 1.
[0083] 2. Use the SnaPShot technology platform to detect the genotype of the SNP site of Scaffold2233:468642 in each sample.
[0084] This method designs primers of different lengths for different mutation sites. After the SNaPshot reaction, the products are separated by electrophoresis, detected by five-color fluorescence, and analyzed by Gene mapper. Multiple SNP sites can be detected in one sequencing reaction. Using SNaPshot for fixed-point sequence analysis, its basic principle follows the dideoxy termination method in direct DNA sequencing, the difference is that there are only different fluorescently labeled ddNTPs in the PCR reaction....
Embodiment 3
[0122] Example 3 A kit for evaluating the content of tea tree epicatechin
[0123] 1. Composition
[0124] Its nucleotide sequence is shown in the primers shown in SEQ ID NO: 2-3, 2×Taq PCR Master Mix, ddH 2 O.
[0125] Wherein, primer F: CAATGCATAATGCTCTACCC (SEQ ID NO: 2);
[0126] Primer R: TGTGTGCGAATCGTTGAAGC (SEQ ID NO: 3).
[0127] 2. How to use
[0128] (1) Extract the total DNA of tea tree shoots by CTAB method, and ensure that the A260 / A280 of each DNA sample is between 1.8 and 2.0, and the concentration is greater than 100 μg / μl;
[0129] (2) PCR amplification
[0130] The PCR system (10μl) is as follows:
[0131] 2×Taq PCR Master Mix 5μl Primer 0.5μl each DNA template 1μl wxya 2 o
3μl
[0132] The PCR amplification procedure is as follows:
[0133]
[0134] (3) Product purification
[0135] The PCR amplification product was subjected to gel electrophoresis, and then recovered and purified using a commercially avai...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com