Ultra-low frequency mutant nucleic acid fragment detection method, library construction method, primer design method and reagent
A technology for designing nucleic acid fragments and primers, which is applied in biochemical equipment and methods, and the determination/inspection of microorganisms, and can solve problems such as affecting the detection limit, loss of cfDNA templates, and loss of templates
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
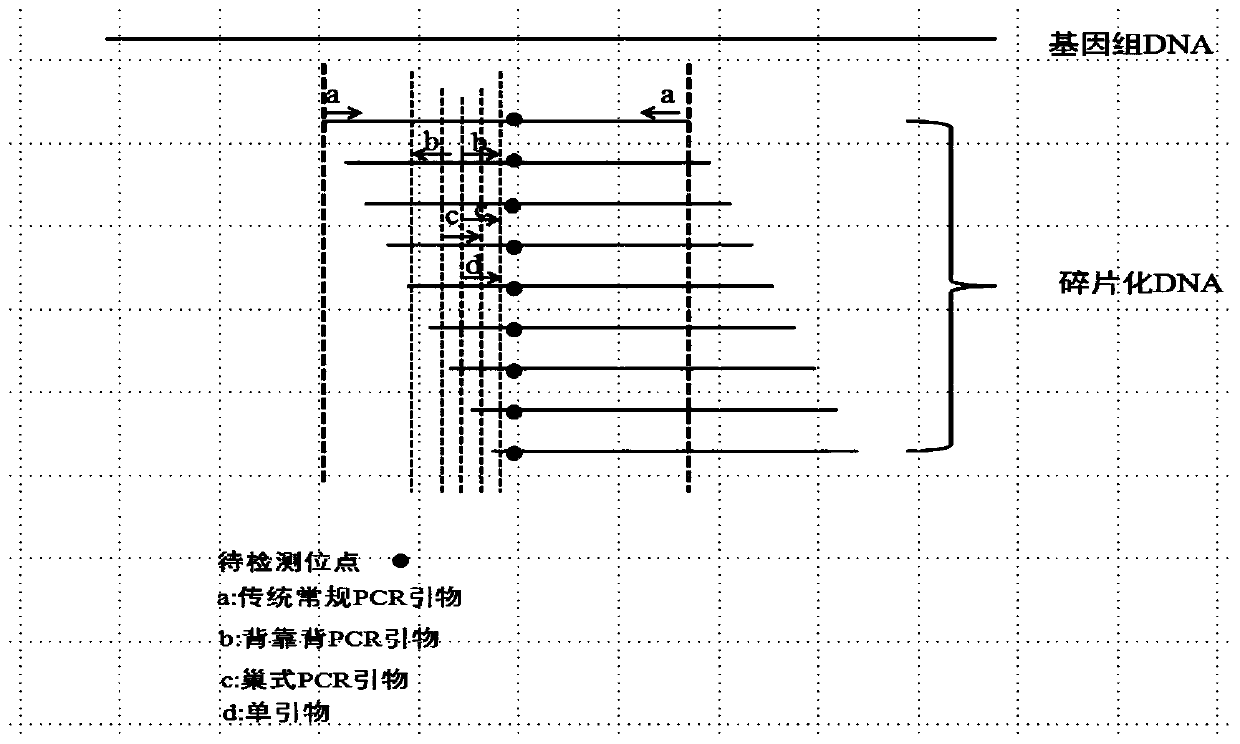
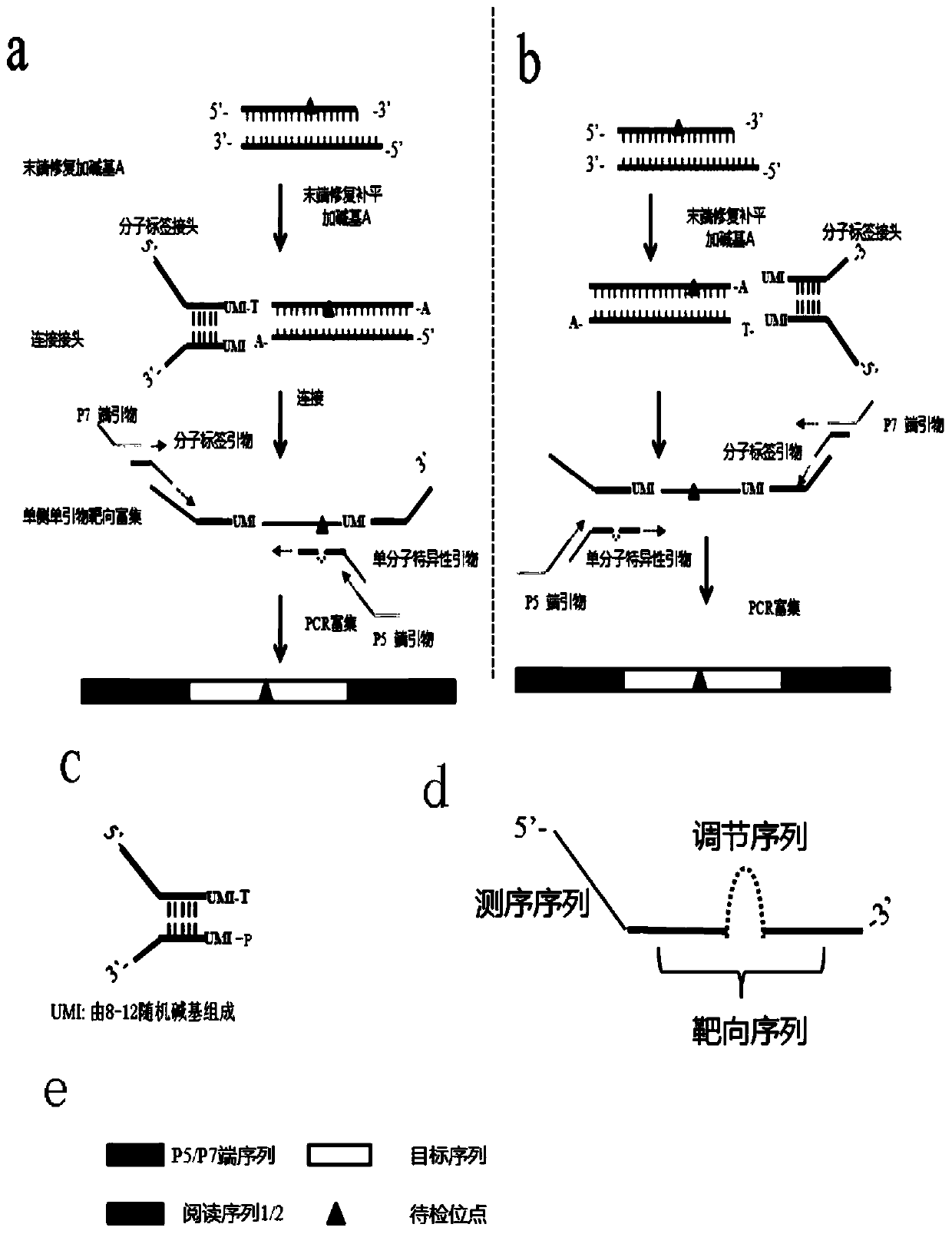
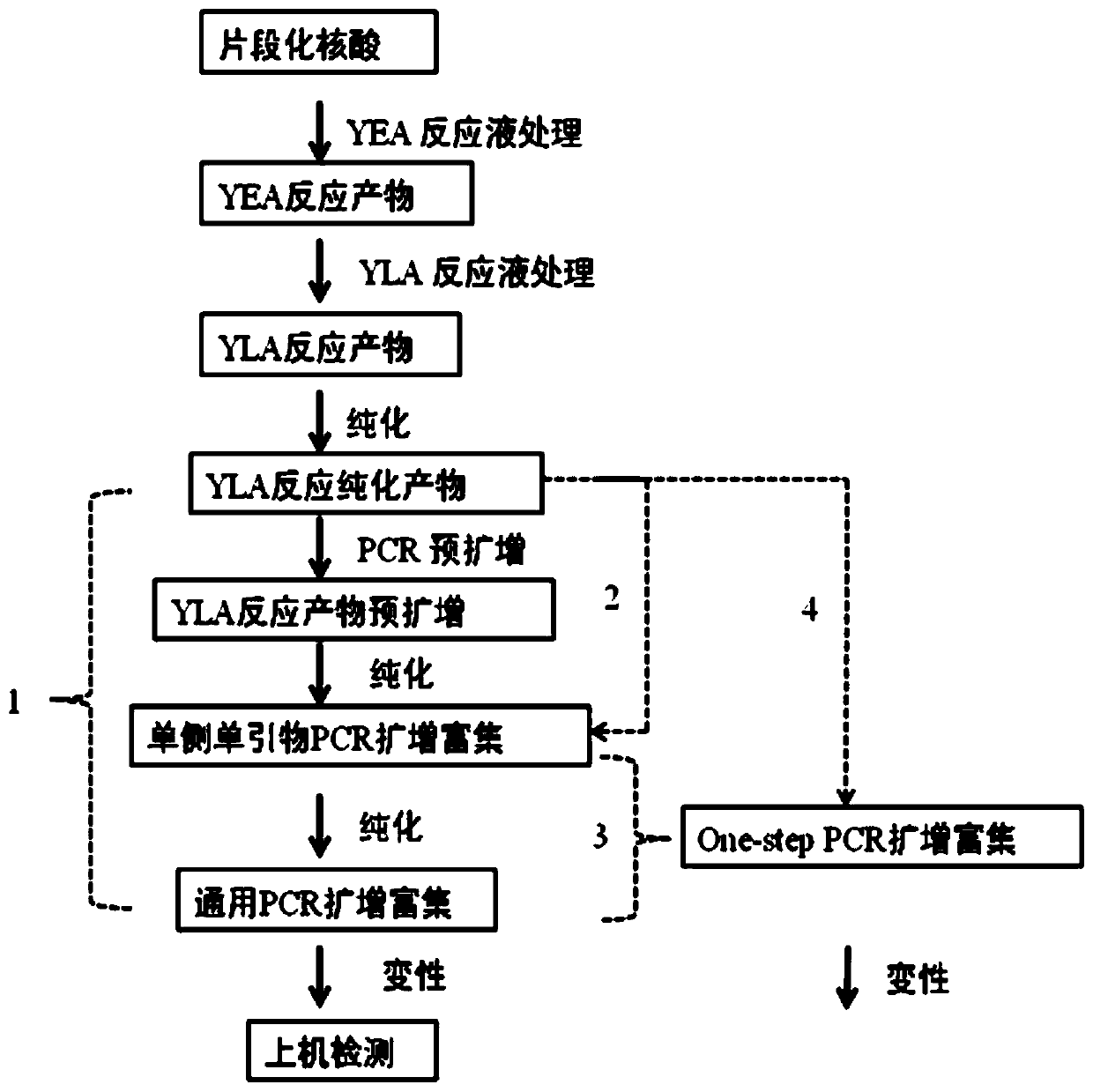
Image
Examples
Embodiment 1
[0112] Example 1 Micro Fragmentation DNA EGFR Gene Mutation Detection
[0113] 1.1 Joint design
[0114] Top-Adaptors:
[0115] 5'-P-ACGTCACGCAGGGGAGAGCCAGGGATGACTAGG-3', see SEQ ID NO.1.
[0116] P indicates phosphorylation modification.
[0117] Bottom-Adapter:
[0118] 5'-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNNNNCCCTGCGTGACGT*Y-3', see SEQ ID NO. 2. * stands for thio modification, Y stands for degenerate base A or T, N stands for random base A or T or G or C, which is used to distinguish and correct errors introduced by PCR and sequencing.
[0119] The above sequences need to be annealed into double strands.
[0120] EGFR gene: (EXON18, EXON19, EXON20, EXON21 below are the 4 exons upstream intron sequence, exon sequence, and downstream intron sequence of the 4 exons of EGFR gene)
[0121] EXON18
[0122] CAAGTGCCGTGTCCTGGCACCCAAGCCCATGCCGTGGCTGCTGGTCCCCCCTGCTGGGCCATGTCTGGCACTGCTTTCCAGCATGGTGAGGGCTGAGGTGACCCTGTCCTCTGTGTTCTTGTCCCCCCAGCTTGTGGAGCCTCTTACACCCAGTGGAGAAGCTC...
Embodiment 2B
[0235] Example 2 BRAF Gene Targeted Sequencing
[0236] 2.1 Joint design
[0237] Top-Adaptors:
[0238] 5'P-ACGTCACGCAGGGNNNNNNNNAGATGTGTATAAGAGACAG-3', see SEQ ID NO.21,
[0239] P indicates phosphorylation modification.
[0240] Bottom-Adapter:
[0241] 5'-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNNNNCCCTGCGTGACGTT-3', see SEQ ID NO. 22. The above sequences need to be annealed into double strands, and N represents a random base A or T or G or C, which is used to distinguish and correct errors introduced by PCR and sequencing.
[0242] The sequence of BRAF exon 11 is as follows:
[0243] AAAACACTTGGTAGACGGGACTCGAGTGATGATTGGGAGATTCCTGATGGGCAGATTACAGTGGGACAAAGAATTG[ G] ATCTGGATCATTT[ GGA] ACAGTCT[ A] CAAGGGAAAGTGGCATG, see SEQ ID NO. 23,
[0244] The sequences of BRAF exon 11 and its upstream and downstream introns are as follows:
[0245] CCTGTATCCCTCTCAGGCATAAGGTAATGTACTTAGGGTGAAACATAAGGTTTTCTTTTTCTGTTTGGCTTGACTTGACTTTTTACTGTTTTTATCAAGAAAACACTTGGTAGACGGGACTCGAGTGATGA...
Embodiment 3
[0345] Example 3 Ultra-low input cf-DNA gene detection one-step library construction
[0346] 3.1 Adapter primer design
[0347] 3.1.1 Joint design
[0348] Top-Adaptors:
[0349] 5'-P-ACGTCACGCAGGGNNNNNNNNAGATGTGTATAAGAGACAG-3', see SEQ ID NO.33,
[0350] P stands for phosphorylation modification.
[0351] Bottom-Adapter:
[0352] 5'-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNNNNCCCTGCGTGACGT*Y-3', see SEQID NO.34, *Y represents the degenerate base A or T modified by sulfo, N represents the random base A or T or G or C, used to distinguish correction PCR and Errors introduced by sequencing.
[0353] The above sequences need to be annealed into double strands.
[0354] Designed for the G12R site of exon 2 of the KRAS gene
[0355] The KRAS exon 2 sequence is as follows:
[0356] ATGACTGAATATAAACTTGTGGTAGTTGGAGCT[ G G]T[GG]CGTAGGCAAGAGTGCCTTGACGATACAGCTAATTCAGAATCATTTTGTGGACGAATATGATCCAACAATAGAG, see SEQ ID NO.35,
[0357] KRAS exon 2 and its upstream and downstream intro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com