Low-initial-quantity plasma free DNA methylation library-building kit and method
A technology of methylation and kits, which is applied in the field of methylation library construction methods and library construction kits, can solve problems such as difficult to effectively meet the requirements of DNA input volume, high DNA input volume, and high amplification cycle number, and achieve The effect of improving the quality of library construction and sequencing, reducing purification steps, and reducing input requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] 1. End repair and A addition
[0081] Use the nucleic acid-containing solution as template DNA for end repair and A addition reactions.
[0082] Reagents: ER / dA enzyme mixture (containing T4 polynucleotide kinase, Taq DNA polymerase, T4 DNA polymerase), 5XER / dA reaction buffer (containing dNTP, Tris-HCl, MgCl 2 , DTT).
[0083] The input amount of template DNA in the 50μl reaction system is at least 1ng, the concentration of T4 polynucleotide kinase is 0.6U / μl, the concentration of Taq DNA polymerase is 0.3U / μl, the concentration of T4 DNA polymerase is 0.5U / μl, and the concentration of dATP is 0.15nmol / μl, dNTP concentration is 0.3nmol / μl, Tris-HCl concentration is 80mM, MgCl 2 The concentration was 15 mM and the concentration of DTT was 7 mM.
[0084] Reaction conditions: incubate at 30°C for 30 minutes, then incubate at 72°C for 30 minutes.
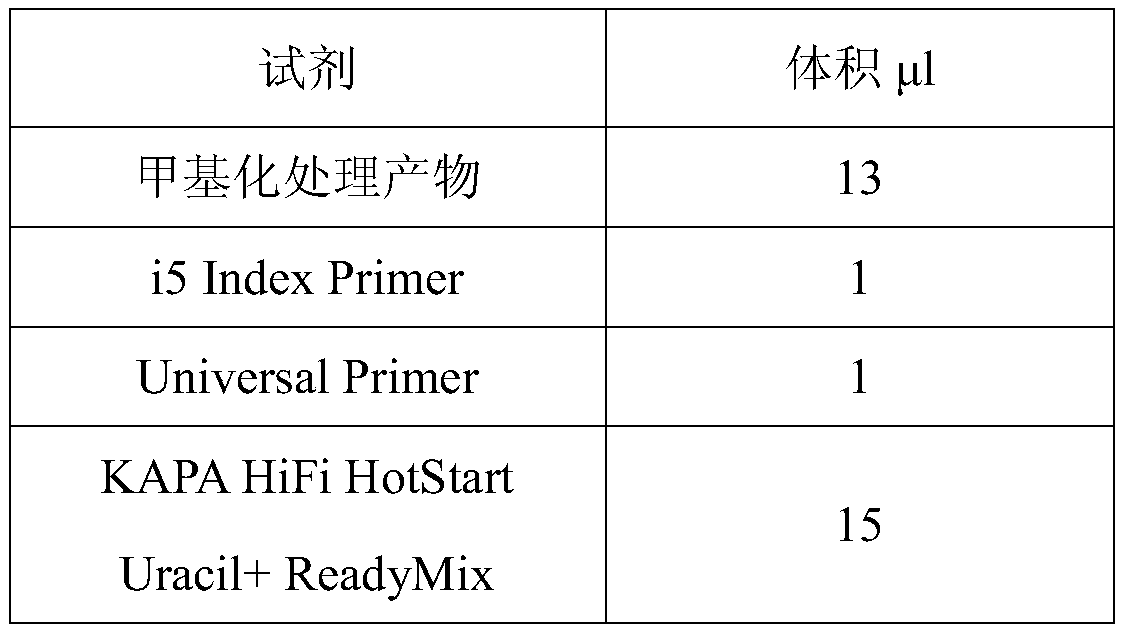
[0085] Specific reaction system:
[0086] Reagent Volume μl template DNA X nuclease free water 32....
Embodiment 2
[0133] The present invention designs a DNA ligase enhancement solution, and tests its effect on the methylation library construction of the present invention.
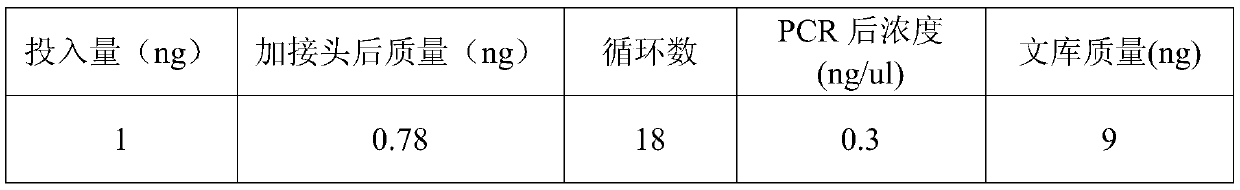
[0134] (1) Test the effect of hexaammine cobalt chloride on the methylation library building of the present invention, taking 10ngDNA input as an example, the results are as follows:
[0135]
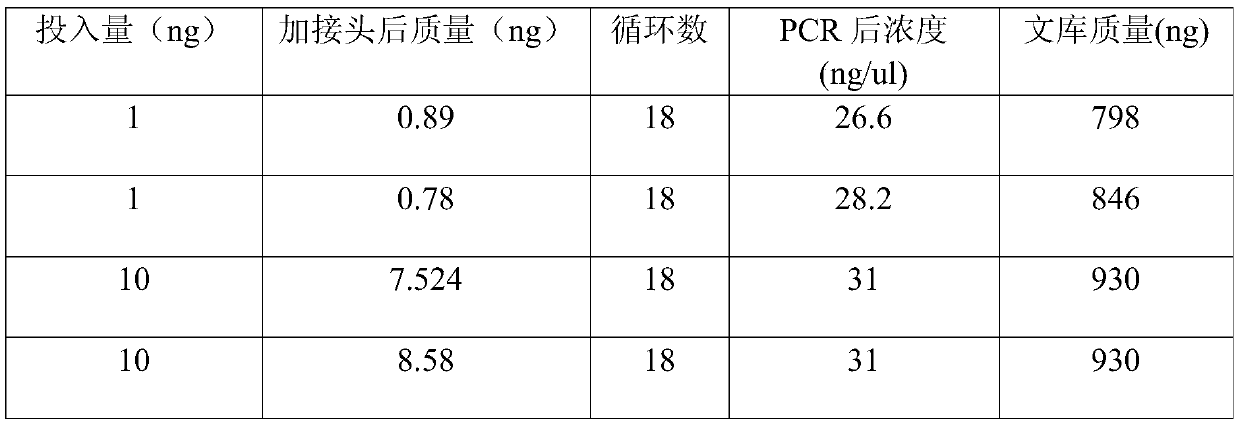
[0136] (2) Test the effect of betaine on methylation library construction of the present invention, taking 10ngDNA input as an example
[0137]
[0138] The results showed that hexaammine cobalt chloride could improve the quality of the library; further addition of betaine could improve the quality of the library to a certain extent.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com