lncRNA lnc12 and application thereof in regulation of development of adventitious roots of poplar
A reaction and sequence technology, applied in the field of plant molecular and molecular biology, can solve the problem of less lncRNA clones, and achieve the effect of delaying the rooting time and reducing the rooting rate.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
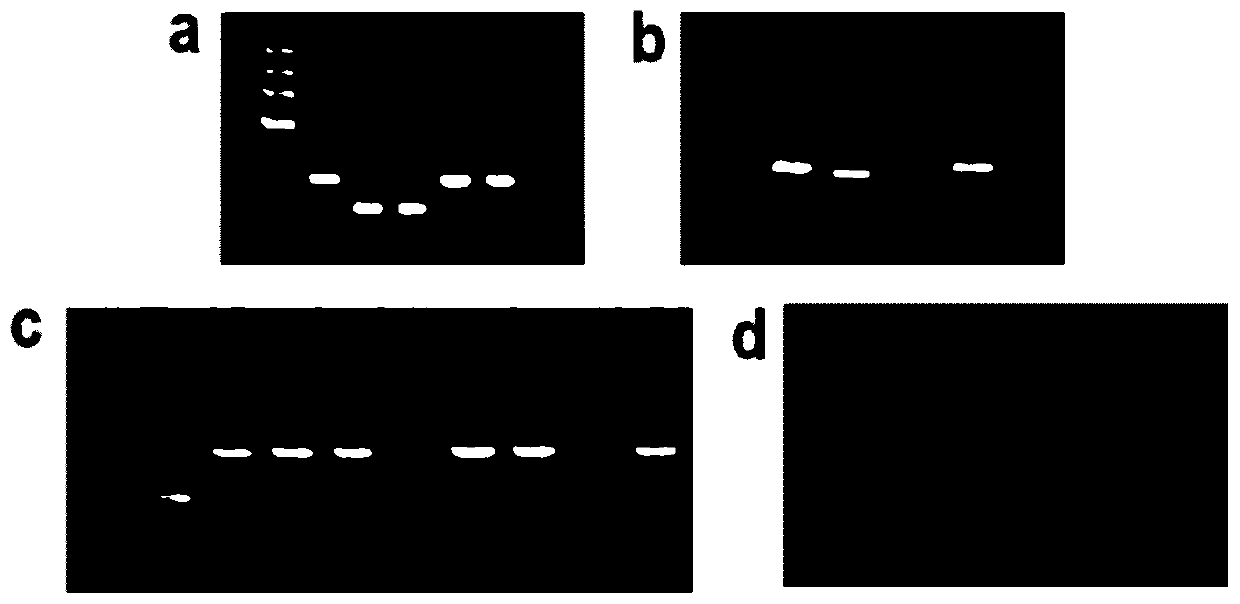
[0030] Example 1 Using RACE technology to clone the full length of lnc12
[0031] 1. RNA extraction
[0032] RNA extraction from the stem (CKS), 1-week-old root (1WR) and 2-week-old root (2WR) of poplar tissue-cultured seedlings of ‘Nanlin 895’.
[0033] 2. Preparation of RACE-Ready cDNA
[0034] RACE-specific primers were designed according to the sequence of lnc12 obtained by high-throughput sequencing. The design principles are: the primer length is 23-28nt, the GC content is 50%-70%, and the annealing temperature T m ≥65°C (Tm≥70°C if using touch down PCR). Use Oligo 6 software to design primers, and the primers used are shown in Table 1:
[0035] Table 1 5'RACE and 3'RACE primers of lnc12
[0036]
[0037] 1) Preparation of Buffer Mix: 4.0 μL of 5×First-Strand Buffer, 0.5 μL of DTT (100 mM), 1.0 μL of dNTP (20 mM), the total volume is 5.5 μL.
[0038] 2) Master Mix preparation: Buffer Mix 5.5μL, RNase Inhibitor 0.5μL, SMART Scribe TM Reverse Transcriptase 2.0 μL...
Embodiment 2
[0072] Example 2 Coding ability prediction and verification of lnc12
[0073] 1. lnc12 coding ability and conservative prediction
[0074]The 5'RACE and 3'RACE sequences of lnc12 were spliced using BioEdit software to obtain the full-length cDNA sequence of lnc12. Primers were designed for the full-length cDNA sequence of spliced lnc12 and verified by sequencing (Table 2). The open reading frame was predicted by NCBI ORFfinder. The coding ability of lnc12 was predicted by CPC software, and the bioinformatics software RegRNA2.0 (http: / / regrna2.mbc.nctu.edu.tw / detection.html) was used to predict whether lnc12 had a ribosome binding site. BlastP was used to perform a similarity search on the possible encoded amino acid sequences of the small open reading frame (short ORF, sORF) of lnc12. The domain of the sORF of lnc12 was predicted using the Pfam database. Search lnc12 in the NONCODE database to predict its conservation. Use Oligo6 to design primers for the predicted sO...
Embodiment 3
[0097] Embodiment 3 poplar protoplast and transformation
[0098] 1. Isolation of poplar protoplasts
[0099] 1) Add in a 10mL centrifuge tube: 200mM MES 500μL, 0.8mol / L mannitol 3.75mL, ddH 2 O 75 μL, 0.2mol / L KCl 500 μL;
[0100] 2) 70°C water bath for 3-5 minutes;
[0101] 3) Add 100 μL cellulase and 25 μL pectinase while hot;
[0102] 4) After 10 minutes in a water bath at 55°C, place on ice and cool to room temperature;
[0103] 5) Add 1mol / L MgCl 2 50 μL, 0.005 g of bovine serum albumin, mix well and filter sterilize.
[0104] 2. Enzymatic hydrolysis of poplar leaves
[0105] 1) Select about 40 days old and well-grown 'Nanlin 895' poplar tissue culture seedling leaves for the preparation of protoplasts;
[0106] 2) Cut the blade into 0.5-1mm wide leaf strips with a blade;
[0107] 3) After the leaves are cut, they are put into the enzyme solution in a stretched state, and cultured in the dark at 28°C for 3 hours;
[0108] 4) Add precooled 5mL W5 solution (2mM M...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com