A primer-probe composition, kit and application for detecting soybean root rot based on recombinase polymerase amplification method
A technology of recombinase polymerase and primer probe, which is applied in the field of genetic engineering, can solve the problems of poor specificity, long cycle and low sensitivity of detection methods, and achieve the effect of convenient operation, fast detection speed and cumbersome operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: Design, screening and validation of specific primer and probe compositions for Pythium ultimum
[0027] In order to obtain the specific primer and probe composition of Pythium ultimum, the present invention firstly downloads Pythium ultimum and more than 20 other Pythium species from the NCBI database based on the bioinformatics analysis tool, using the commonly used ITS sequence as the detection target Through multiple sequence alignment analysis, specific primer compositions were designed in the regions with obvious sequence differences between Pythium ultimum and other Pythium, and 3 groups of primer compositions were preliminarily selected from the designed primer compositions. Composition 1: F1 (TTCACGATGTATGGAGACGCTGCATTTAGTTG), R1 (TCCTCCGCTTATTGATATGCTTAAGTTCAG); primer composition 2: F2 (TTTCAGCAGTGGATGTCTAGGCTCGCACATCG), R2 (GATTCTCGATCGAAAAAACGAACGCAACCAT); For the above three groups of primer compositions, the common PCR method was used to screen ...
Embodiment 2
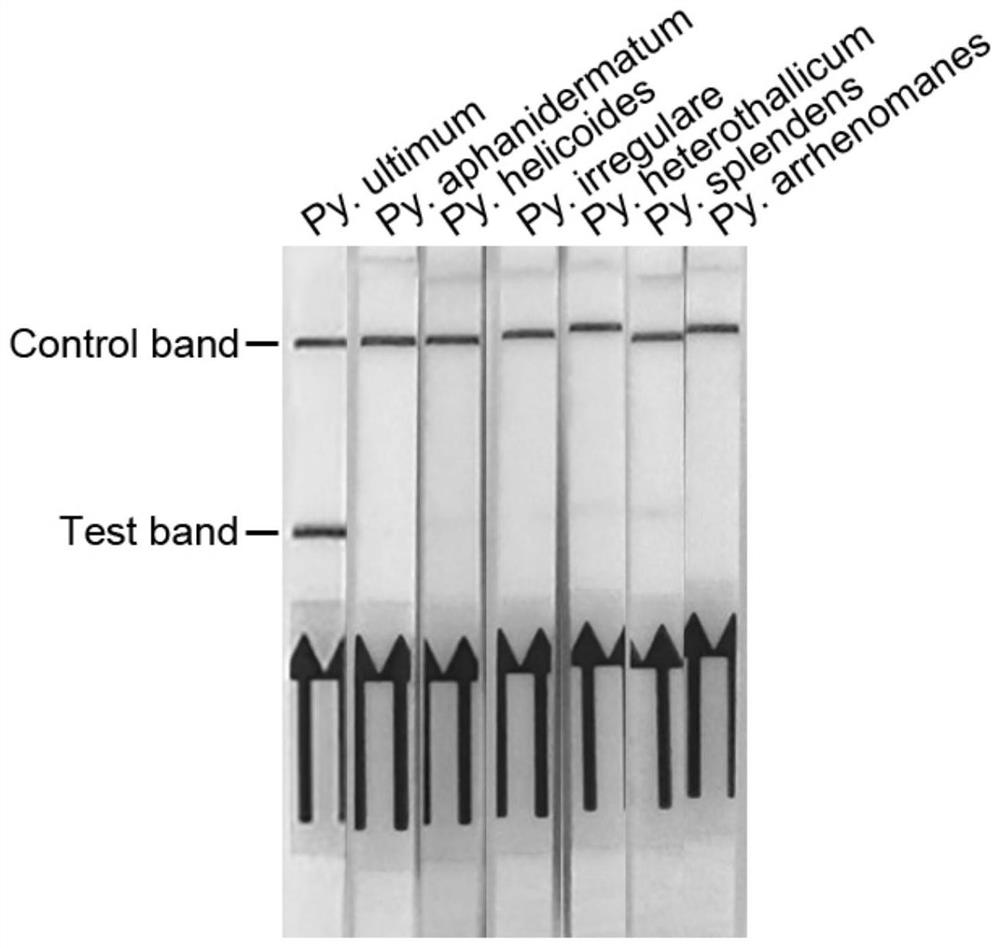
[0028]Example 2: Specificity test of primer and probe composition of Pythium ultimum
[0029] Primer and probe combination for detection of Pythium ultimum: the forward primer has the nucleotide sequence of PuRPA-F as shown in SEQ ID No. 1 and the reverse primer has the PuRPA as shown in SEQ ID No. 2 -R nucleotide sequence, the probe has the PuProbe nucleotide sequence as shown in SEQ ID No.3.
[0030] Forward primer PuRPA-F: TTCACGATGTATGGAGACGCTGCATTTAGTTG
[0031] Reverse primer PuRPA-R: Biotin-TCCTCCGCTTATTGATATGCTTAAGTTCAG
[0032] Probe PuProbe: FAM-TATCATTGTCAATTGCAAGATTGTGTATGG-THF-ATCTCAATTGGACCT-C3 spacer
[0033] The above-mentioned primer and probe combination was used to make a kit. The concentration and dosage of each reagent in the kit were: 2.1 μL 10 μM forward primer PuRPA-F, 2.1 μL 10 μM reverse primer PuRPA-R, 0.6 μL 10 μM of the probe PuProbe, 29.5 μL of Rehydration Buffer, 12.2 μL of DEPC-treated water, 2.5 μL of 280 mM MgAC and dry enzyme powder were m...
Embodiment 3
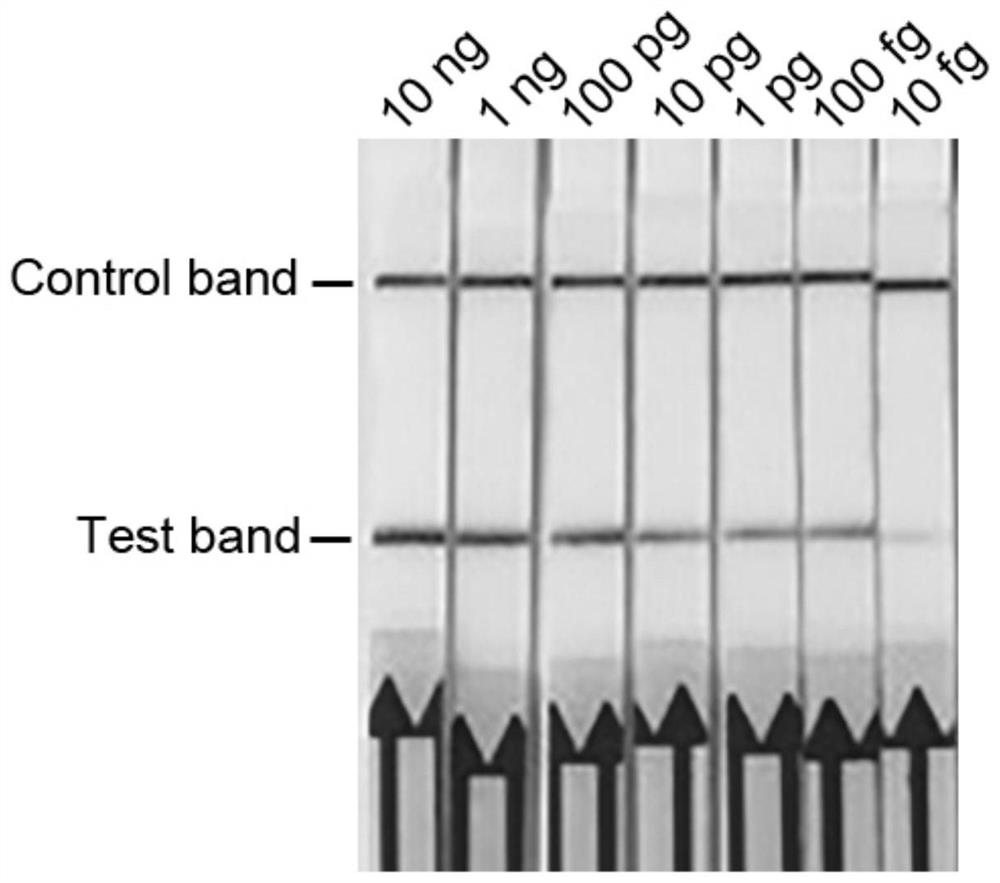
[0036] Example 3: Sensitivity test for detection of Pythium ultimum recombinase polymerase
[0037] In order to determine the sensitivity of the recombinase polymerase detection method, the genomic DNA of Pythium ultimum was measured with a spectrophotometer and then diluted by 10-fold gradient. The DNA concentration range was set to 1ng-10fg. 1 μL was used as a template, DEPC-treated water was used as a negative control, and 49 μL of the kit solution was added to carry out the recombinase polymerase amplification reaction. The reaction program was: incubation at 39°C for 20 min. After the reaction is completed, use lateral flow chromatography test strips to detect the amplification products, and judge the detection results according to the color change of the indicator band of the detection line. The results show that: DNA concentration in the range of 1ng-100fg can observe the strip at the detection line of the test strip, the DNA concentration is 10fg and the negative contr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com