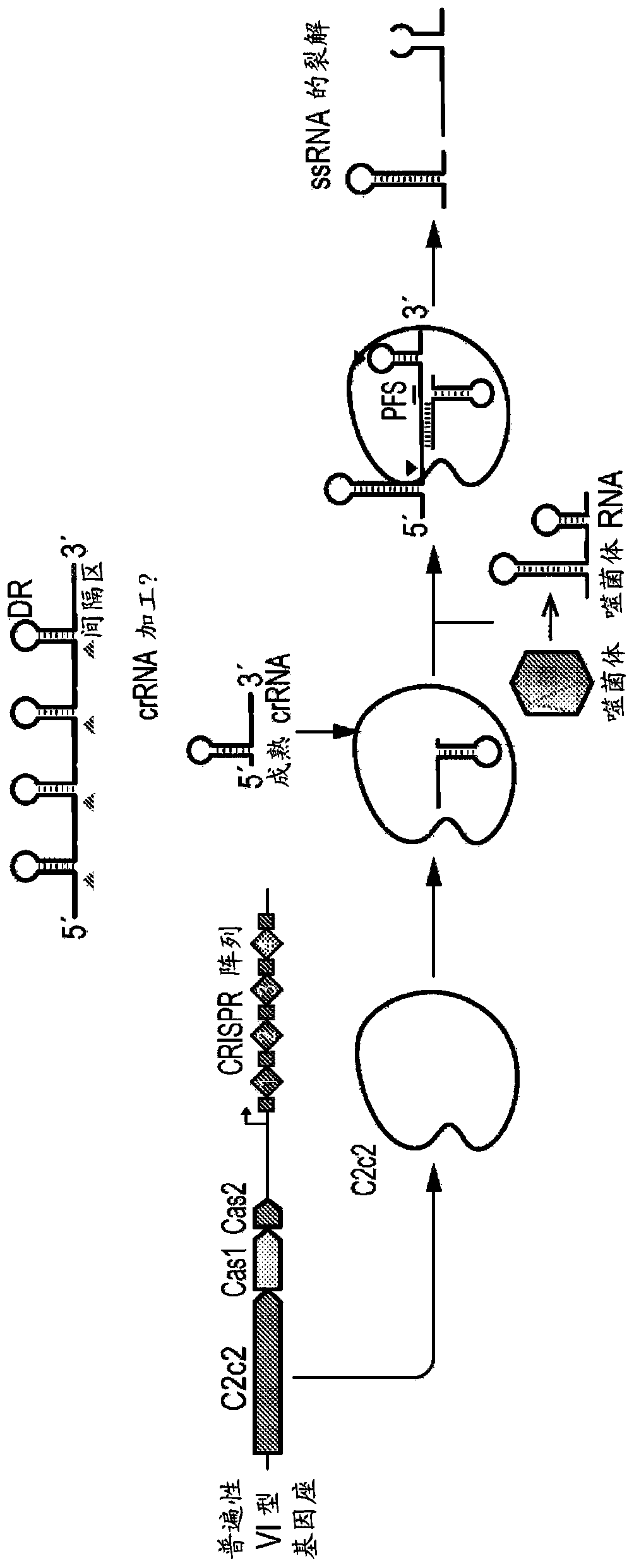
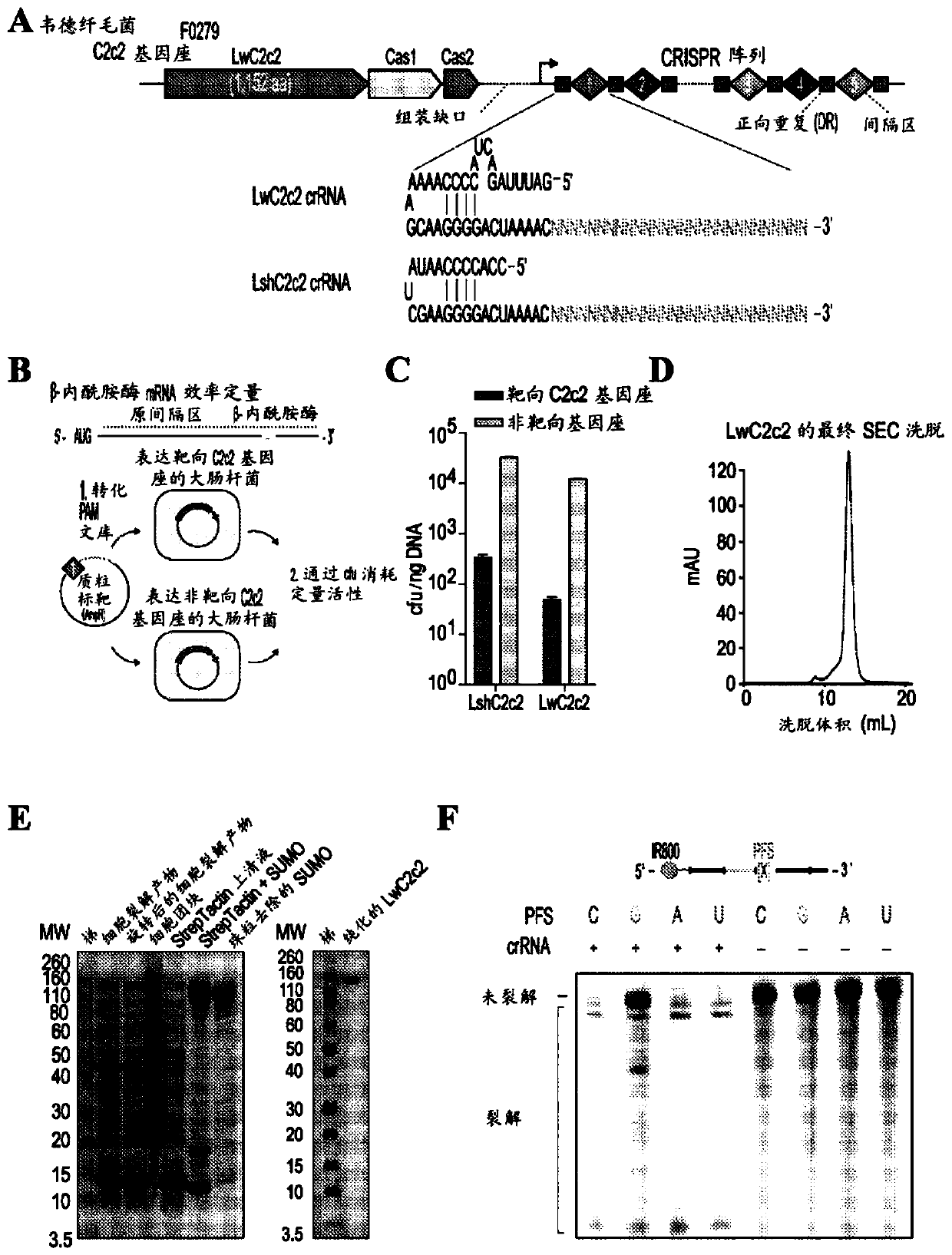
Crispr effector system based diagnostics for virus detection
A detection system, virus technology, applied in genetic engineering, microbial determination/inspection, DNA/RNA fragments, etc., can solve problems such as expensive, low-sensitivity applications, limited availability, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1- 1
[0654] Example 1 - General Protocol
[0655] There are two ways to perform DNA and RNA diagnostic tests for C2c2. This protocol can also be used with protein detection variants after delivery of detection aptamers. The first is a two-step reaction in which amplification and C2c2 detection are done independently. The second is where everything is combined in one reaction and this is called a two-step reaction. It is important to keep in mind that amplification may not be necessary for higher concentration samples, so it is good to have a separate C2c2 protocol that does not have amplification embedded in it.
[0656] Table 10. CRISPR Effectors Only - No Amplification:
[0657] component Volume (μL) Protein (final 44nM) 2 crRNA (final 12nM) 1 Background target (100ng total) 1 target RNA (variable) 1 RNA sensor probe (125nM) 4 MgCl 2 (final 6mM)
[0658] The reaction buffer was: 40 mM Tris-HCl, 60 mM NaCl, pH 7.3
[0659] This ...
Embodiment 2
[0672] Example 2 - Highly sensitive and specific detection of C2C2-mediated DNA and RNA from C. wiederii
[0673] Rapid, inexpensive, and sensitive nucleic acid tests can aid in point-of-care pathogen detection, genotyping, and disease surveillance. The RNA-guided RNA-targeting CRISPR effector Cas13a (previously known as C2c2) exhibits a "side effect" of promiscuous RNase activity upon target recognition. We combined the collateral effects of Cas13a with isothermal amplification to establish a CRISPR-based diagnostic (CRISPR-Dx), providing rapid DNA or RNA detection with attomolar sensitivity and single-base mismatch specificity. We used this Cas13a-based molecular detection platform, called SHERLOCK (Specific High Sensitivity Enzymatic Reporter UnLOCKing), to detect specific strains of Zika and Dengue viruses, differentiate pathogenic bacteria, genotyping human DNA, and identifying DNA mutations in cell-free tumors. In addition, SHERLOCK reagents can be lyophilized for cold...
Embodiment 3-C2c2
[0791] Example 3 - C2c2 prevents infection and reduces replication of lymphocytic choriomeningitis (LCMV)
[0792] like Figure 67 As indicated in and listed in Table 20 below, guide RNAs were designed to bind to various regions of the LCMV genome.
[0793] Table 20
[0794] gRNA annotation gRNA-seq SEQ ID NO L1 ctcacgaagaaagttgtgcaaccaaaca 624 L2 tcttcaatctggttggtaatggatattt 625 L3 gccaatttgttagtgtcctctataaatt 626 L4 tatctcacagaccctatttgattttgcc 627 L5 aaattcttcattaaattcaccatttttg 628 L6 tatagtttaaacataactctctcaattc 629 S1 atccaaaaagcctaggatccccggtgcg 630 S2 agaatgtcaagttgtattggatggttat 631 S3 aaagcagccttgttgtagtcaattagtc 632 S4 tgatctctttcttctttttgtcccttac 633 S5 tctctttcttctttttgtcccttactat 634 S6 caatcaaatgcctaggatccactgtgcg 635
[0795] Lipofectamine 2000 was used to transfect into 293FT cells a plasmid expressing C. wiede (Lw) C2c2 / Cas13a fused to msfGFP with nuclear ex...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com