Novel coronavirus N-S dominant epitope fusion protein, preparation method, application, expression protein, microorganism, application and kit
A dominant epitope, fusion protein technology, applied in the field of genetic engineering, can solve the problems of difficult nucleic acid sampling, long detection time, missed detection of infected persons, etc., to achieve excellent sensitivity and specificity, improved detection sensitivity, and reduced missed detection rate. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1 N protein dominant epitope segment antigen screening and determination
[0027] According to the full-length amino acid sequence of the new coronavirus N protein published in Genebank, the amino acid sequence of the N protein was analyzed with the B-cell epitope analysis and signal peptide analysis functions of BIOSUN software, and the antigenicity was selected according to the strength of hydrophobicity and signal peptide. Good 20-220aa segment (denoted as N1), 230-300aa segment (denoted as N2) and 335-419aa segment (denoted as N3) are used as segment antigens for screening the dominant epitope of N protein in this example .
[0028] The gene sequences of segments N1 to N3 are:
[0029] The N1 gene sequence is:
[0030] PSDSTGSNQNGERSGARSKQRRPQGLPNNTASWFTALTQHGKEDLKFPRGQGVPINTNSSPDDQIGYYRRATRRIRGGDGKMKDLSPRWYFYYLGTGPEAGLPYGANKDGIIWVATEGALNTPKDHIGTRNPANNAAIVLQLPQGTTLPKGFYAEGSRGGSQASSRSSSRNSSRNSSPRMNSTPNGGSSRGTALP
[0031] The N2 gene sequence is:
[0032] ...
Embodiment 2
[0049] Example 2 S protein dominant epitope segment antigen screening and determination
[0050] According to the full-length amino acid sequence of the new coronavirus S protein published in Genebank, the amino acid sequence of the S protein was analyzed by using the BIOSUN software B-cell epitope analysis and signal peptide analysis functions, and combined with the distribution of functional domains, selected 65- The 290aa segment (denoted as S1), the 340-510aa segment (denoted as S2) and the 520-681aa segment (denoted as S3) three dominant epitope segment antigens are used to screen the S protein dominant epitope in this example segment antigens.
[0051] The S1 gene sequence is:
[0052]fhaihvsgtngtkrfdnpvlpfndgvyfasteksniirgwifgttldsktqsllivnnatnvvikvcefqfcndpflgvyyhknnkswmesefrvyssannctfeyvsqpflmdlegkqgnfknlrefvfknidgyfkiyskhtpinlvrdlpqgfsaleplvdlpiginitrfqtllalhrsyltpgdsssgwtagaaayyvgylqprtfllkynengtitdavd
[0053] The S2 gene sequence is:
[0054] evfnatrfasvyawnrkr...
Embodiment 3
[0065] Example 3 Preparation of N-S dominant epitope fusion protein of novel coronavirus
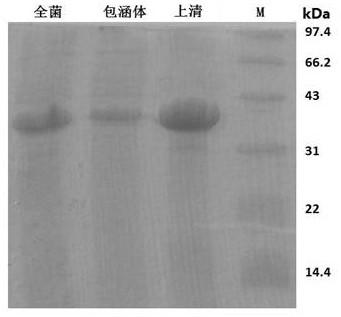
[0066] In this example, the coding genes of the N1 dominant epitope segment antigen and the S3 dominant epitope segment antigen were linked together for fusion expression using the homologous enzyme method with complementary sticky ends to obtain the N-S dominant epitope fusion protein of the new coronavirus, Its amino acid sequence is as follows:
[0067] PSDSTGSNQNGERSGARSKQRRPQGLPNNTASWFTALTQHGKEDLKFPRGQGVPINTNSSPDDQIGYYRRATRRIRGGDGKMKDLSPRWYFYYLGTGPEAGLPYGANKDGIIWVATEGALNTPKDHIGTRNPANNAAIVLQLPQGTTLPKGFYAEGSRGGSQASSRSSSRSRNSSRNSTPGSSAPATVCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTGSNVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNSP
[0068] The nucleic acid sequence of the above-mentioned novel coronavirus N-S dominant epitope fusion protein is as follows:
[0069]CCGAGCGATTCTACCGGCTCCAACCAGAATGGTGAACGTTCGGGCGCGCGCAGCAAACA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com