Copy number variation detection method and device
A copy number variation and detection method technology, applied in the fields of instrumentation, genomics, proteomics, etc., can solve the problem of low sensitivity of mutation detection, and achieve the effect of improving sensitivity, high coverage, and accurate resolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
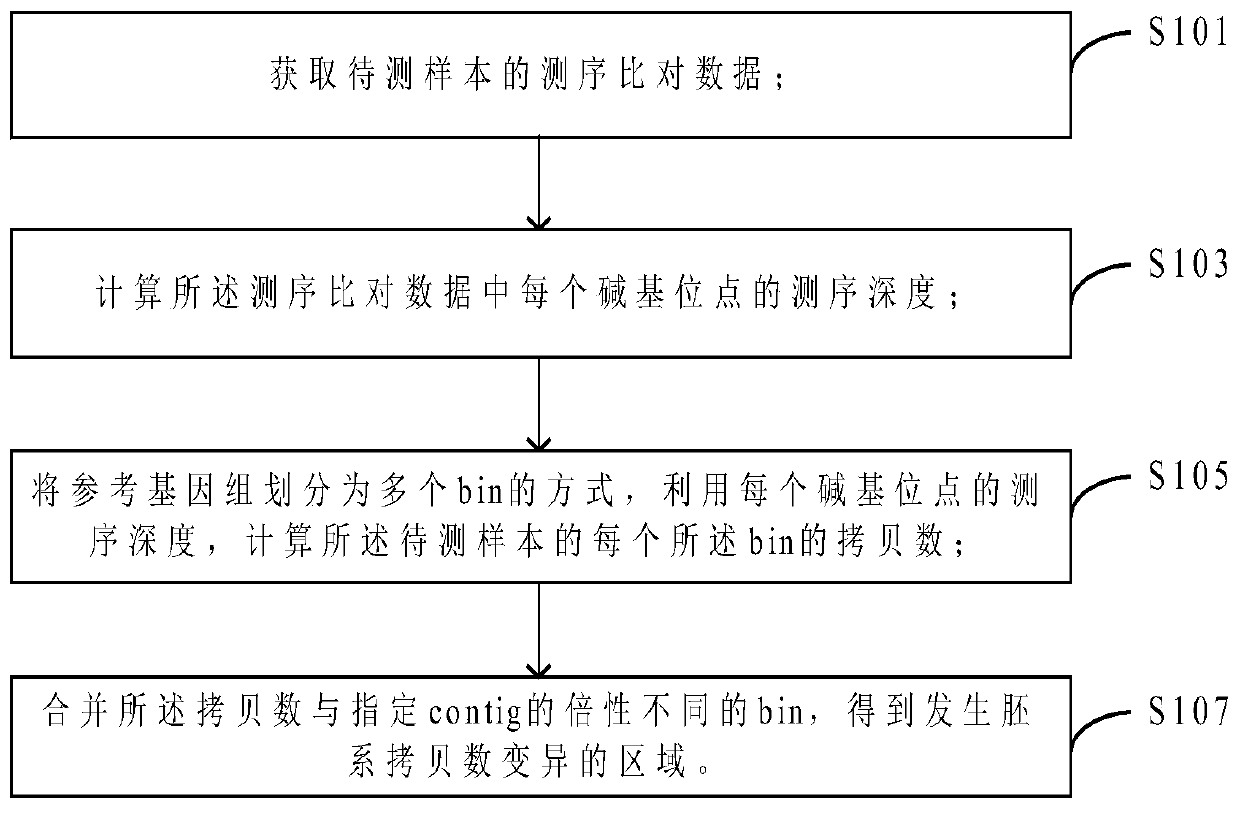
[0033] In this embodiment, a method for detecting copy number variation is provided, such as figure 1 As shown, the detection method includes:
[0034]Step S101, obtaining the sequencing comparison data of the sample to be tested;
[0035] Step S103, calculating the sequencing depth of each base site in the sequencing alignment data;
[0036] Step S104, divide the reference genome into multiple bins, and calculate the copy number of each bin of the sample to be tested by using the sequencing depth of each base site;
[0037] Step S107, merging the bins whose copy number is different from the ploidy of the specified contig to obtain the region where the germline copy number variation occurs.
[0038] The detection method of the above-mentioned copy number variation obtains the ploidy (ie copy number) of each bin by using a bin-based method, and then compares it with the ploidy of the specified contig to be divided into a plurality of different bins, However, the differential...
Embodiment 2
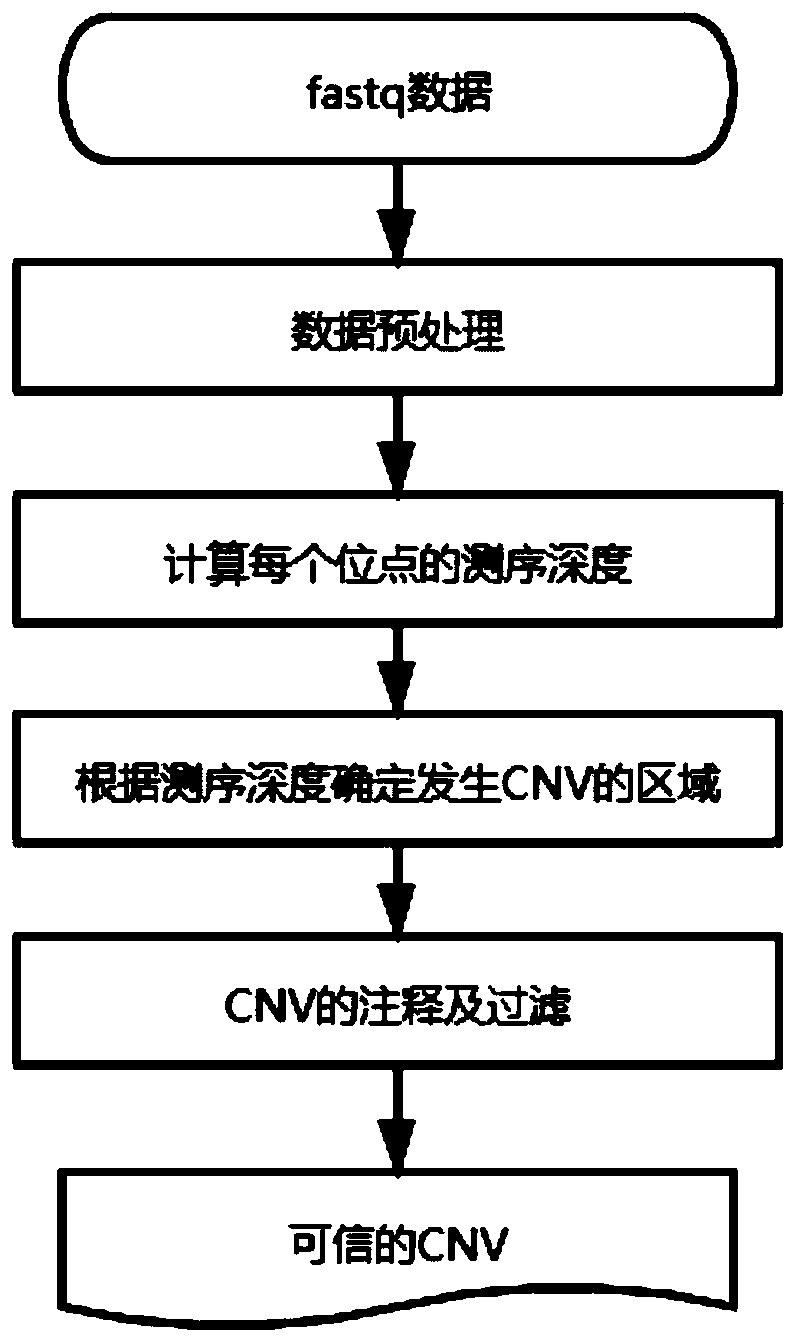
[0052] This embodiment provides a specific detection method for copy number variation, such as figure 2 shown, including the following steps:
[0053] 1. Data preprocessing
[0054] Input the result file of NGS data in fastq format, and generate the sequence alignment result in bam format.
[0055] 1) Preprocess the original off-machine data to remove reads containing adapters and low quality.
[0056] 2) Compare the processed raw data with the reference genome to obtain a comparison result file in bam format;
[0057] 3) Remove the sequence alignment results outside the range of the capture chip;
[0058] 4) remove the repeated reads in the comparison result file, and obtain a bam file that does not contain the repeated comparison result;
[0059] 2. Calculate the sequencing depth of each base site
[0060] Calculate the sequencing depth of each base site in the specified region in the bam file, and obtain a file in tsv format.
[0061] 3. Determine the CNV region acco...
Embodiment 3
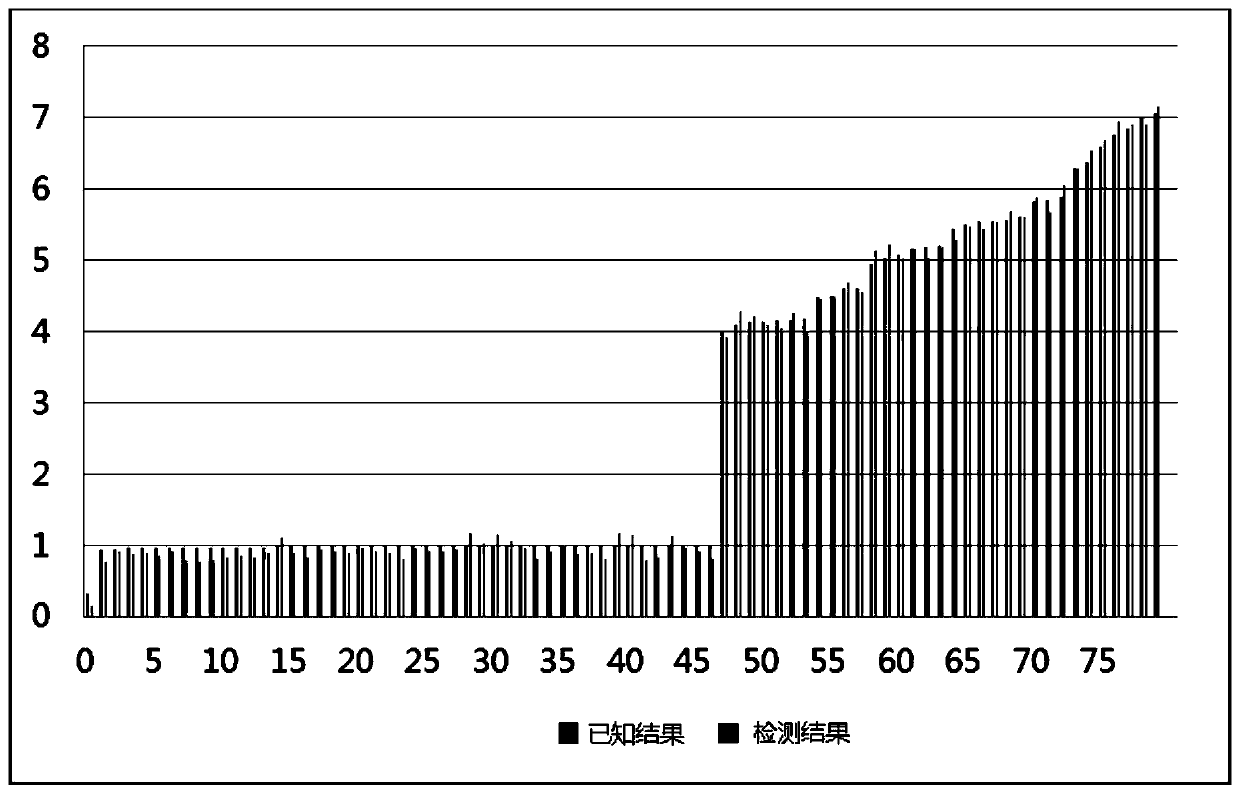
[0069] Using the method of Example 2, the positive samples with known copy number variation were detected, and the results were as follows: image 3 As shown, the horizontal axis is the sample, the vertical axis is the copy number, the dark gray indicates the positive sample result, and the light gray indicates the detection result. It can be seen from the detection results that the detection results are consistent with the known results. It can be seen that the detection method of the present application can not only detect all the mutation sites, but also has high detection accuracy.
[0070] It should be noted that for the foregoing method embodiments, for the sake of simple description, they are expressed as a series of action combinations, but those skilled in the art should know that the present invention is not limited by the described action sequence. Because of the present invention, certain steps may be performed in other orders or simultaneously. Secondly, those sk...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com