Method for regulating plant salt tolerance and salt tolerance related protein
A salt-tolerance and salt-tolerance technology, applied in the field of regulating plant salt-tolerance and salt-tolerance-related proteins, can solve problems such as lack of understanding
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] Embodiment 1, the cloning of the rice OsPRR73 coding gene related to salt tolerance
[0084] The inventors of the present invention isolated and cloned a rice gene OsPRR73 related to salt tolerance from rice Nipponbare, as shown in Sequence 1 of the Sequence Listing, and named its encoded protein OsPRR73 protein, as shown in Sequence 2 of the Sequence Listing.
[0085] The rice Nipponbare total RNA was extracted and reverse-transcribed into cDNA, and PCR amplification was performed with primers F:ATGGGTAGCGCCTGCGAAGC and R:TTATCTGTCTTCGTCTTGGC. The PCR reaction program was: pre-denaturation at 94°C for 2 min; denaturation at 94°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 30 s, and 35 cycles; extension at 72°C for 5 min. The PCR products were subjected to Sanger sequencing. Sequencing results show that the nucleotide sequence of the PCR amplification product is sequence 1 in the sequence listing, its coding sequence is the 1st-2304th nucleotide of sequ...
Embodiment 2
[0086] Embodiment 2. OsPRR73 deletion mutant rice construction
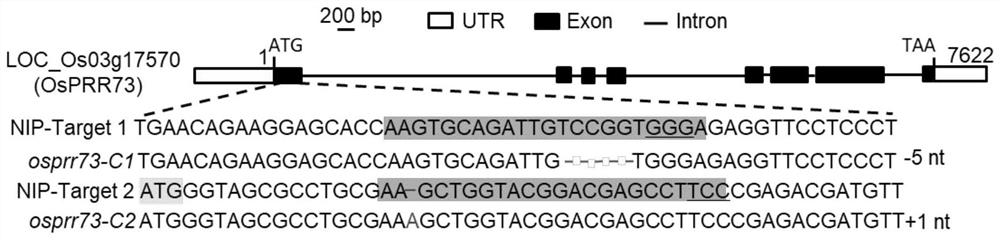
[0087] 1. Construction of vectors and recombinant bacteria
[0088]Use the DNA sequence shown in Sequence 1 to screen for suitable targets on the E-CRISPR website (http: / / www.e-crisp.org / E-CRISP / designcrispr.html). Combining the scores and target positions, ATGGGTAGCGCCTGCGAAGCTGG located on the first exon of OsPRR73 was selected as one of the CRISPR / Cas9 targets. Synthetic primer sequences F: ggcATGGGTAGCGCCTGCGAAGC and R: aaacGCTTCGCAGGCGCTACCCA, according to the literature (Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R ,Chen S,Wang Z,Chen Y,Guo J,Chen L,Zhao X,Dong Z,Liu Y-G(2015)A Robust CRISPR / Cas9 System for Convenient,High-Efficiency Multiplex Genome Editing in Monocot and DicotPlants.Mol Plant 8: 1274-1284) to construct the pCRISPR / Cas9 vector system to the vector pCRISPR / Cas9-OsPRR73. The vector pCRISPR / Cas9-OsPRR73 was introduced into Agrobacterium EHA105 comp...
Embodiment 3
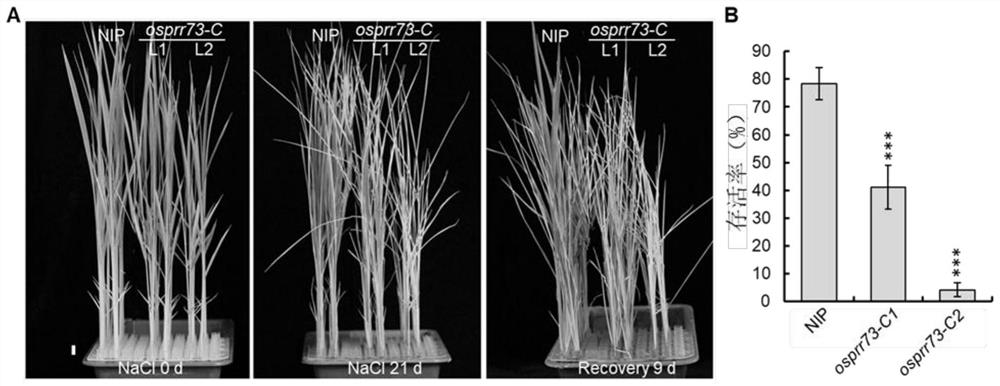
[0093] Example 3 Identification of transgenic rice CRISPR / Cas9-OsPRR73 T0 generation plants
[0094] DNA was extracted from the leaves of the T0 generation of the rice OsPRR73 mutant, and the extracted DNA was used as a template to perform PCR amplification with OsPRR73-specific primers. The primer sequences were F: ACGTTGGGCGTATGATGCA; R: TCACCATGCTATGAAGCT. The PCR reaction program was: pre-denaturation at 94°C for 2 min; denaturation at 94°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 30 s, and 35 cycles; extension at 72°C for 5 min. The PCR product was subjected to Sanger sequencing to check whether the target site was mutated.
[0095] Wherein, the extraction of leaf DNA was carried out as follows: rice leaves were put into a 1.5 mL centrifuge tube, steel balls were added and ground into powder with a grinder. Add 500 μL of extraction buffer to the centrifuge tube and shake to mix. Add an equal volume of chloroform and phenol (1:1) mixture, shake and mix...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com