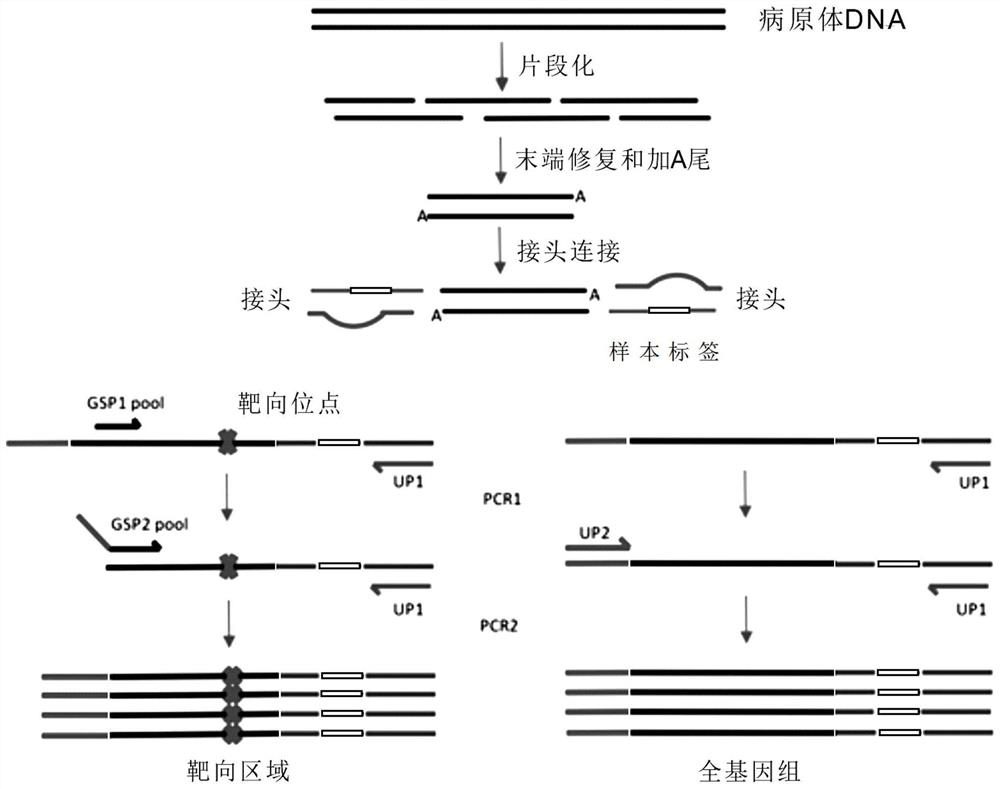
Whole genome combined targeted amplification library building method, reagent and pathogen detection method
A whole genome and gene technology, applied in the field of sequencing detection, can solve the problems of missed detection of pathogens, insufficient sensitivity, high bacterial content, etc., and achieve the effect of reducing detection cost, improving detection efficiency, and increasing detection rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] The detection of embodiment 1 pathogen
[0057] The components in the following Table 1 were formulated into a reference product according to a certain ratio, and then Yanhuang (YH) genome: Escherichia coli genome: reference product genome were mixed according to the ratio of 9:90:1, and simulated samples were prepared for subsequent experiments.
[0058] Table 1
[0059] pathogen nature Porcine herpesvirus (Suid herpesvirus) dsDNA Fowlpox virus dsDNA Klebsiella oxytoca Gram-negative bacteria (-) Enterococcus faecalis Gram-positive bacteria (+) Candida glabrata fungus
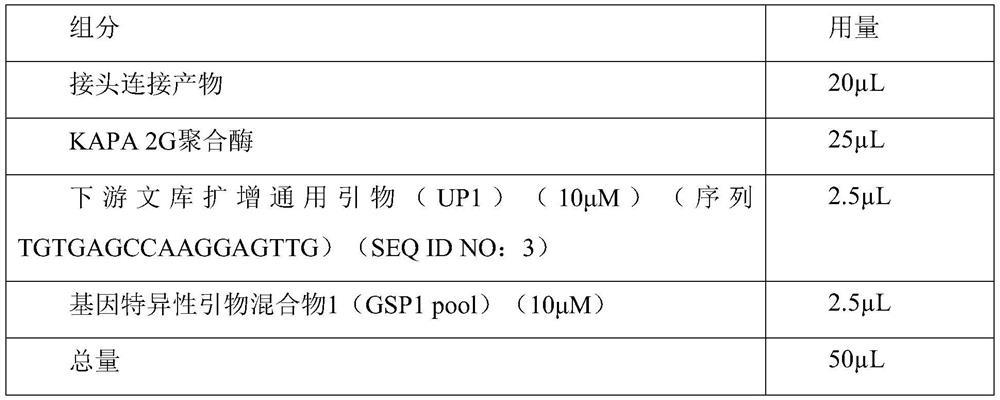
[0060] (1) DNA fragmentation:
[0061] Configure the reaction system as shown in Table 2, and incubate at 37°C for 25 minutes.
[0062] Table 2
[0063] components Dosage 10×reaction buffer 2μL fragmentase (NEB) 0.5μL mock sample 10 μL water 7.5μL Total 20 μL
[0064] (2) Make up and add A reaction
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com