Artificially designed plasmid pM5500 and application thereof in preparation of DNA marker
A plasmid, artificial technology, applied in DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problems of complicated preparation process and insufficiently compact plasmid structure, and achieve high amplification efficiency, stability and stability. Amplification efficiency, easy-to-estimate effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
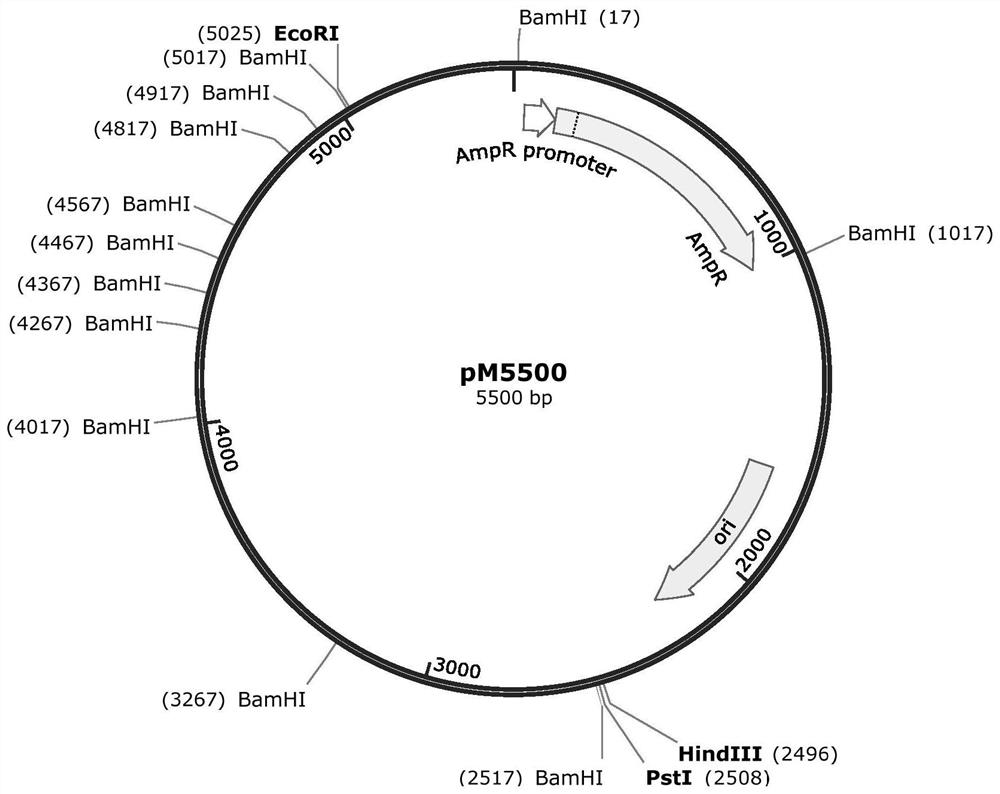
[0024] like figure 1 , this embodiment provides a plasmid pM5500 for the preparation of medium molecular weight DNA markers, its sequence is shown in SEQ ID NO: 1, including high-copy replication origin sequence and ampicillin resistance gene coding sequence, and various artificially designed Combined DNA fragments. The plasmid contains 12 homogeneous restriction endonuclease recognition sites within a limited length, and can produce 1500, 1000, 750, 500, 250 and 100bp common DNA fragments in six medium-molecular-weight DNA markers by single enzyme digestion, And the mass ratio of various fragments is 3:2:3:1:1:1.
Embodiment 2
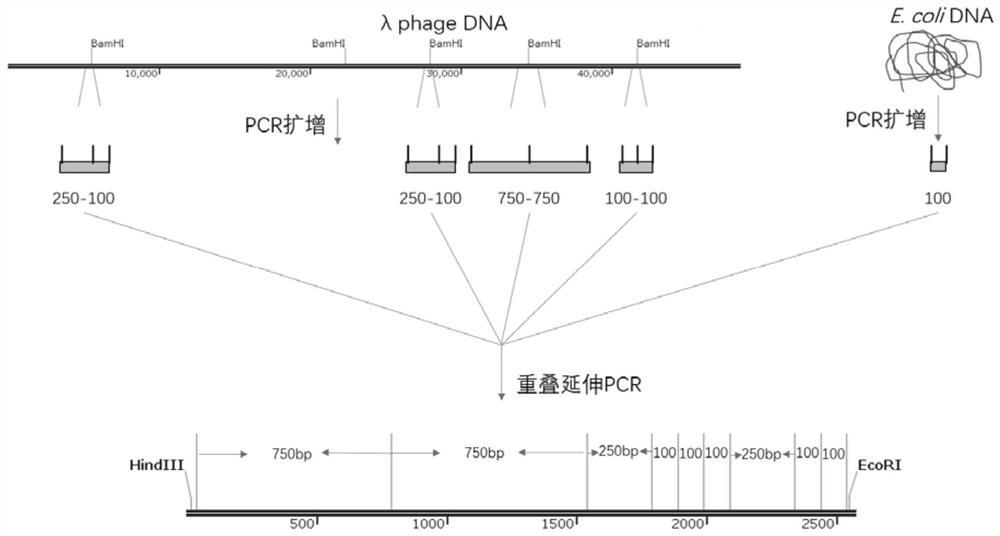
[0026] This example provides a method for constructing pM5500 and similar plasmids: select a suitable infrastructure plasmid, such as pUC19, pBR322, LITMUS28i, etc., introduce restriction enzyme sites into the vector infrastructure, and make the distance between the cutting points meet expected. Additional length fragments are subsequently introduced at the multiple cloning site of the vector. Finally, the plasmid contained one fragment of 1500bp, one fragment of 1000bp, two fragments of 750bp, one fragment of 500bp, two fragments of 250bp and five fragments of 100bp. These fragments can be derived from known plasmids, or amplified from DNA of other species, or obtained by chemical synthesis. Fragments as desired in the present invention were amplified from phage lambda and E. coli genomes. The designed plasmid is 5500bp in length and contains 12 recognition sites for a specific restriction endonuclease (12 BamH I recognition sites in this example). The total length of the ...
Embodiment 3
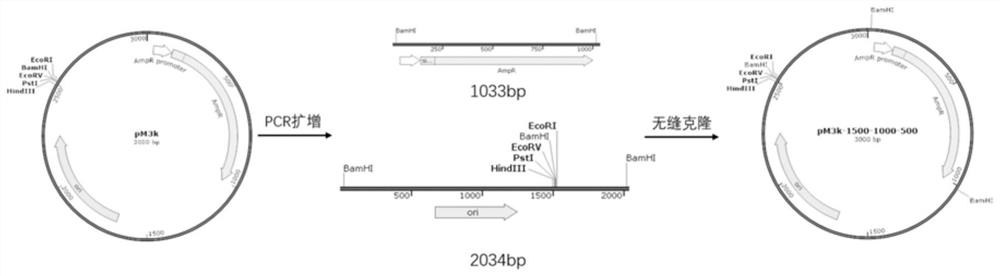
[0028] This embodiment provides a construction of pM5500 and similar vectors, and the construction of pM5000 is divided into three steps:
[0029] Step 1: Transformation of the basic plasmid backbone. The construction starts with a plasmid called pM3K, which is transformed based on the LITMUS28i plasmid, with a size of 3kb and a BamH I cutting site. like figure 2 As shown, a BamH I cutting point is introduced in the upstream and downstream of the ampicillin resistance gene of the backbone plasmid pM3K, so that the modified pM3K contains 3 BamH I cutting points, and three fragments of 1500, 1000, and 500 bp are produced after enzyme digestion. When introducing the BamH I cutting point, the pM3K plasmid was first used as a template to amplify, and the primers contained the BamH I recognition sequence, that is, the upstream and downstream 15-20nt sequences, and two DNA fragments with a length of 1033bp and 2034bp were obtained respectively. Then use Novizym's ClonExpress II On...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com